Avalekander Week 11: Difference between revisions
From OpenWetWare
Jump to navigationJump to search
Avalekander (talk | contribs) →Methods: added how i found concentration value |
Avalekander (talk | contribs) →Acknowledgements: acknowledged Leanne |
||
| Line 42: | Line 42: | ||
*I would like to acknowledge my homework partner, [[User:EdwardRyanTalatala|Edward]] with whom I worked with to complete the assignment. | *I would like to acknowledge my homework partner, [[User:EdwardRyanTalatala|Edward]] with whom I worked with to complete the assignment. | ||
*I would also like to acknowledge [[User:Kam D. Dahlquist|Dr. Dahlquist]], as well as [[User:Ben G. Fitzpatrick|Dr. Fitzpatrick]]. | *Additionally, I would like to acknowledge [[User:Leannekuwahara| Leanne]] with whom I communicated with via text message when I was confused about the MatLab script versus dynamic files and where the parameter values should be plugged in. | ||
*I would also like to acknowledge [[User:Kam D. Dahlquist|Dr. Dahlquist]], as well as [[User:Ben G. Fitzpatrick|Dr. Fitzpatrick]] for their help throughout the assignment. | |||
Except for what is noted above, this individual journal entry was completed by me and not copied from another source.[[User:Avalekander|Avalekander]] ([[User talk:Avalekander|talk]]) 19 | Except for what is noted above, this individual journal entry was completed by me and not copied from another source. [[User:Avalekander|Avalekander]] ([[User talk:Avalekander|talk]]) 14:19, 9 April 2019 (PDT) | ||
==References== | ==References== | ||
Revision as of 21:19, 9 April 2019
Purpose
- The purpose of this assignment is again familiarizing ourselves with MatLab and using it to perform calculations on the ongoing study being done with data collected from the Dahlquist Lab.
Methods
- This file was read and used for background information: Chemostat Modeling Assignment.
- The MATLAB files named chemostat_script.m and chemostat_dynamics.m were used to simulate a chemostat and compare the computations to a steady state outcome.
- The following is the zip file used: here
- The parameter values used were:
- q = 0.10 (1/hr)
- u = 5 (g/L)
- E = 1.5
- r = 0.8 (1/hr)
- K = 8 (g)
- The parameter values were plugged into the following equations to formulate the steady states of cell biomass and nutrient mass:
- y = qK/(r-q)
- x = (u-y)/E
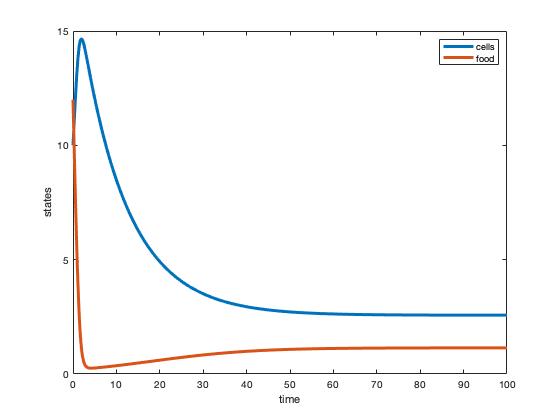
- Assuming a 2 liter chemostat, the steady state concentrations of cells and nutrients were then calculated by dividing the previous values found for x and y by 2 L.
- The system dynamics were simulated using the MATLAB files and the parameters of (1). The following questions were then answered:
- Do the graphs show the system going to steady state?
- Do the steady states match your (1) calculations?
- The graphs wre uploaded to our individual journals.
- BONUS: can you get two y-axes, with the second one to the right of the picture like in the journal articles you’ve read?
Results
- Parameters were plugged into y = qK/(r-q) and x = (u-y)/E to solve for cell biomass and nutrient mass (g) at equilibrium
- Nutrient mass- y=(0.1(8))/(0.8-0.1)=1.14 g
- Cell biomass- x=(5-1.14)/1.5 = 2.6 g
- Concenration in a 2L chemostat:
- Nutrient mass = 1.14g/2L = 0.55 g/L
- Cell biomass = 2.6g/L = 1.3 g/L
- MatLab Simulation:
Conclusion
Acknowledgements
- I would like to acknowledge my homework partner, Edward with whom I worked with to complete the assignment.
- Additionally, I would like to acknowledge Leanne with whom I communicated with via text message when I was confused about the MatLab script versus dynamic files and where the parameter values should be plugged in.
- I would also like to acknowledge Dr. Dahlquist, as well as Dr. Fitzpatrick for their help throughout the assignment.
Except for what is noted above, this individual journal entry was completed by me and not copied from another source. Avalekander (talk) 14:19, 9 April 2019 (PDT)
References
- Dahlquist, K. & Fitpatrick, B. (2019). "BIOL388/S19: Week 11" Biomathematical Modeling, Loyola Marymount University. Accessed from:Week 11 Assignment Page
Biology 388 Assignments
- Template: Ava Lekander
- User Page: Ava Lekander
- Journal Entries:
- Assignment Pages:
- Class Journal Pages:
- Biology 388 Home Page: BIOL 388 Class Page