BacFITBase Review
From OpenWetWare
Jump to navigationJump to search
Purpose
The purpose of this assignment is to analyze the BacFITBase, a database that characterizes the pathogenesis of bacterial proteins. Additionally, the purpose of this assignment is to learn more about bacterial proteins and possible antibiotic targets.
Methods/Results
Database Evaluation
- First, we read the article about the database from the Nucleic Acids Research journal, and then we went online to the database itself
- After browsing through online databases, we decided to analyze BacFITBase: a database to assess the relevance of bacterial genes during host infection (https://academic.oup.com/nar/article/48/D1/D511/5608989)
General information about the database
- What is the name of the database? (link to the home page)-Kam
- The name of the database is BacFitBase
- What type (or types) of database is it? -Owen
- BacFITBase is primarily a protein sequence database; however, this database could also be characterized as a 3-D protein structure database and a model organism database. This is because the primary search results are proteins and their respective amino acid sequences; however, all UniProt proteins automatically have their 3-D structure generated via ProViz, and the pathogenesis of the proteins can be filtered through multiple host species such as mice, chickens, rabbits, and cows. (http://www.tartaglialab.com/bacfitbase/display?query=bacfit0019077)
- What biological information (type of data) does it contain? (sequence, structure, model organism, or specialty [what?])
- BacFITBase contains a lot of information, and all of the information revolves around bacterial pathogens and their ability to infect host species (http://www.tartaglialab.com/bacfitbase/display?query=bacfit0019077).
- Bacterial Genes and the specific proteins they encode
- Bacterial Protein Sequences
- Bacterial Protein 3-D Structures
- Specific Pathogen Species
- Specific Host Species
- Infection Fitness Scores for the Bacterial Gene/Protein
- BacFITBase contains a lot of information, and all of the information revolves around bacterial pathogens and their ability to infect host species (http://www.tartaglialab.com/bacfitbase/display?query=bacfit0019077).
- What type of data source does it have?
- BacFITBase has gathered its data from primary sources, "15 different studies where transposon mutagenesis was performed"(http://www.tartaglialab.com/bacfitbase/about).
- BacFITBase is a manually curated database, so it is curated by humans (http://www.tartaglialab.com/bacfitbase/about).
- In-house staff curates the database.
- What individual or organization maintains the database? Is it public or private? -Kam
- This database is freely available to the public, and is maintained by Javier Macho, Benjamin Lang, and Gian Gaetano Tartaglia
- large national or multinational entity or small lab group?
- Those who maintain this database are a small lab group
- What is their funding source(s)? -Owen
- BacFITBAse is "funded by the Spanish Ministerio de Ciencia, Innovación y Universidades (SAF2015-72518-EXP, SAF2017-82158-R and RYC-2012-09999) and a Research Grant 2016 by the European Society of Clinical Microbiology and Infectious Diseases (ESCMID)," and it is additionally funded by "the European Union’s Horizon 2020 research and innovation programme under the Marie Sklodowska-Curie grant agreement No 793135" (http://www.tartaglialab.com/bacfitbase/about).
Scientific quality of the database
- Does the content appear to completely cover its content domain?-Kam
- The content appears to cover its content domain entirely, as it includes information regarding the protein sequence of bacteria, as well as their structures. Furthermore, this database also collects data from transposon mutagenesis experiments that are publicly available, in order to standardize all fitness scores for mutant genes.
- How many records does the database contain?
- This database contains 90,000 entries, including information from 15 pathogenic bacteria among 5 host vertebrates, across 10 various tissues.
- What claims do the database owners make about coverage in the corresponding paper?
- They claim that BacFitBase "applies a standardized reprocessing to published data to enable assessment of how important genes from 15 pathogenic bacteria are to infection of five vertebrate hosts" (https://academic.oup.com/nar/article/48/D1/D1/5695332)
- What species are covered in the database? (If it is a very long list, summarize.) -Owen
- There are 15 bacterial species in the BacFITBase. To summarize, the bacterial species range from the common E. coli and Salmonella bacteria to relatively less known vibrio parahaemolyticus. Despite the wide range of bacterial species, they all share one thing in common: they are involved in the manifestation of disease/infection. For example, BacFITBase contains data on porphyromonas gingivalis, a bacterial species that I am interested in due to its association with periodontitis).
- Acinetobacter baumannii ATCC 17978 (http://www.tartaglialab.com/bacfitbase/browse?pathogen=400667)
- Campylobacter jejuni subsp. jejuni 81-176 (http://www.tartaglialab.com/bacfitbase/browse?pathogen=354242)
- Escherichia coli CFT073 (http://www.tartaglialab.com/bacfitbase/browse?pathogen=199310)
- Escherichia coli M12 (http://www.tartaglialab.com/bacfitbase/browse?pathogen=1392858)
- Escherichia coli O157:H7 str. EDL933 (http://www.tartaglialab.com/bacfitbase/browse?pathogen=155864)
- Haemophilus influenzae Rd KW20 (http://www.tartaglialab.com/bacfitbase/browse?pathogen=71421)
- Klebsiella pneumoniae subsp. pneumoniae ATCC 43816 KPPR1 (http://www.tartaglialab.com/bacfitbase/browse?pathogen=1308539)
- Mycobacterium avium subsp. paratuberculosis K10 (http://www.tartaglialab.com/bacfitbase/browse?pathogen=262316)
- Porphyromonas gingivalis ATCC 33277 (http://www.tartaglialab.com/bacfitbase/browse?pathogen=431947)
- Salmonella enterica Serovar Typhimurium SL1344 (http://www.tartaglialab.com/bacfitbase/browse?pathogen=216597)
- Salmonella enterica Serovar Typhimurium ST4 74 (http://www.tartaglialab.com/bacfitbase/browse?pathogen=909946)
- Serratia marcescens Strain UMH9 (http://www.tartaglialab.com/bacfitbase/browse?pathogen=615)
- Streptococcus pyogenes M1 5448 (http://www.tartaglialab.com/bacfitbase/browse?pathogen=301447)
- Vibrio cholerae O1 biovar El Tor str. N16961 (http://www.tartaglialab.com/bacfitbase/browse?pathogen=243277)
- Vibrio parahaemolyticus RIMD 2210633 (http://www.tartaglialab.com/bacfitbase/browse?pathogen=223926)
- There is a short list of host species in BacFITBase (http://www.tartaglialab.com/bacfitbase/)
- Cow (Bos taurus)
- Chicken (Gallus gallus)
- Mouse (Mus musculus)
- Rabbit (Oryctolagus cuniculus)
- Pig (Sus scrofa)
- There are 15 bacterial species in the BacFITBase. To summarize, the bacterial species range from the common E. coli and Salmonella bacteria to relatively less known vibrio parahaemolyticus. Despite the wide range of bacterial species, they all share one thing in common: they are involved in the manifestation of disease/infection. For example, BacFITBase contains data on porphyromonas gingivalis, a bacterial species that I am interested in due to its association with periodontitis).
- Is the database content useful? I.e., what biological questions can it be used to answer?- Kam
- The database content is useful, as it provides a large number of fitness scores from transposon mutagenesis experiments. These experiments contribute greatly to the identification of genes that are fundamental to infect a specific host organism. By standardizing all of these fitness scores, this database will contribute greatly to the development of new antimicrobial therapies.
- Is the database content timely? -Owen
- The BacFITBase is extremely timely. The COVID-19 pandemic has shown just how vulnerable the human population is to diseases and infections caused by microbes, and the pandemic has also illustrated the need for information on the pathogenesis of diseases. Although the COVID-19 pandemic was caused by SARS-CoV-2, a virus, and the BacFITBase is only a database for bacterial pathogens, the need for this critical information still remains. Additionally,"the development of new antimicrobial therapies relies heavily on our understanding of the mechanisms of bacterial infection. Therefore, it is crucial to understand how bacterial infection develops in vivo and which bacterial genes are required to infect a host" (http://www.tartaglialab.com/bacfitbase/about).
- Is there a need in the scientific community for such a database at this time?
- There is definitely a need for the BacFITBase in the scientific community at this time. Perhaps the biggest reason is simply because it puts all of the bacterial pathogenesis data that is available in one place. Furthermore, the need for novel therapeutics and antibiotics has never been greater, so the BacFITBase is very important for the scientific community to have right now (http://www.tartaglialab.com/bacfitbase/about).
- Is the content covered by other databases already?
- There are databases that have similar information, but none as specific and extensive as BacFITBase (http://www.tartaglialab.com/bacfitbase/about).
- How current is the database?- Kam
- This database is new, it is from 2019.
- When did the database first go online?
- This database first went online in March of 2019
- How often is the database updated?
- This database seems as if it will be continually updated, however, it does not say how often.
- When was the last update?
- January 8th, 2020
General utility of the database to the scientific community
- Are there links to other databases? Which ones? -Owen
- There are links to other databases (http://www.tartaglialab.com/bacfitbase/about)
- Basic Local Alignment Search Tool (BLAST)
- UniProt
- ProViz
- BioPortal
- There are links to other databases (http://www.tartaglialab.com/bacfitbase/about)
- Is it convenient to browse the data?- Kam
- Yes, the layout of the page is very simple, with a large search bar in the middle of the screen, making it seem very user-friendly.
- Is it convenient to download the data? -Owen
- Yes, the data is easy to download and it is user-friendly.
- In what file formats are the data provided?
- What type of files, indicated by the file extension (e.g., .txt, .xml., etc.)?
- Are they standard or non-standard formats? (i.e., are they following an approved standard for that type of data)?
- These formats are standard formats
- Evaluate the “user-friendliness” of the database: can a naive user quickly navigate the website and gather useful information? -Kam
- A naive user would be able to quickly navigate the website, by either searching for their particular bacterial strain of interest on the home page in the large search bar, or clicking the about section at the top of the page to read about the contents of the database. Furthermore, if they are having trouble understanding the database, they could always click on the 'tutorial' tab at the top of the page.
- Is the website well-organized?
- The website is well-organized, as it includes the main home page with the search bar, and tabs at the top, sectioned as such: BLAST, Browse, Download, Tutorial, and About. It does not look too busy like other databases either.
- Does it have a help section or tutorial?
- Yes, the 'tutorial' tab is at the top left of the page.
- Are the search options sensible?
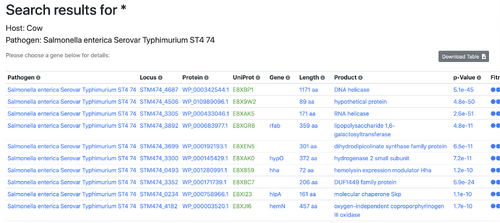
- Yes, the options include various host species such as the cow, pig, chicken, mouse, or rabbit, along with a variety of pathogens to choose from.
- Run a sample query. Do the results make sense?
- Access: Is there a license agreement or any restrictions on access to the database? -Owen
- There is no restriction on access BacFITBase; however, they do provide a link to the Centre for Genomic Regulation's Legal and Privacy Policy (http://www.tartaglialab.com/bacfitbase/about).
Summary judgment
- Would you direct a colleague unfamiliar with the field to use it?-Kam
- Yes, I would recommend this database to someone who is unfamiliar with the field, because it has a tutorial section that could guide them step by step. Furthermore, it could inform them about the various pathogens that cause disease in certain hosts, which is important knowledge to have.
- Is this a professional or "hobby" database? The "hobby" analogy means that it was that person's hobby to make the database. It could mean that it is limited in scope, done by one or a few persons, and/or seems amateur.
- This is a professional database because it has quite a bit of funding from both the Spanish Ministerio de Ciencia and the European Union’s Horizon 2020 research and innovation programme.
- Finally, please share why you chose this database in the first place, i.e., why did it interest you? Did it live up to the expectations you had when you chose it? -Owen
- This database initially caught my eye because as I was scrolling through the specific species of bacteria that the database contained, I noticed that it contained information on P. gingivalis. Being someone who is pursuing a career in dentistry, this specific bacteria interests me due to their ability to form oral biofilms. I believe the database lived up to my expectations, but I believe there is much more room for expansion.
Conclusion
The BacFITBase is a protein sequence database that allows researchers to assess the relevance of bacterial genes during host infection. The database is user-friendly, informational, and professional, and I would recommend it to a colleague performing research on bacterial protein pathogenecity.
Acknowledgments
- My homeork partner, Kam Taghizadeh, and I contacted each other to divide the work evenly
- We copied and modified the procedures and question found on the BIOL368/F20 week 8 page
- Except for what is noted above, this individual journal entry was completed by me and not copied from another source
Owen R. Dailey (talk) 16:48, 28 October 2020 (PDT)
References
- Introduction to the 2020 NAR Database Issue: [https://academic.oup.com/nar/article/48/D1/D1/5695332 Rigden, D. J., & Fernández, X. M. (2020). The 27th annual Nucleic Acids Research database issue and molecular biology database collection. Nucleic Acids Research, 48(D1), D1-D8. doi:
- Javier Macho Rendón, Benjamin Lang, Gian Gaetano Tartaglia, Marc Torrent Burgas, BacFITBase: a database to assess the relevance of bacterial genes during host infection, Nucleic Acids Research, Volume 48, Issue D1, 08 January 2020, Pages D511–D516, https://doi.org/10.1093/nar/gkz931
- OpenWetWare. (2020). BIOL368/F20:Week 8. Retrieved Octiber 24, 2020, from https://openwetware.org/wiki/BIOL368/F20:Week_8
Template
User Page
Assignments
Individual Journals
The D614G Research Group Week 12
The D614G Research Group Week 14
Class Journals
Links to Weekly Assignments
Links to Individual Journal Assignments
- Kam Taghizadeh
- Kam Taghizadeh Week 2
- Kam Taghizadeh Week 3
- Kam Taghizadeh Week 4
- Kam Taghizadeh Week 5
- Kam Taghizadeh Week 6
- Kam Taghizadeh Week 7
- BacFITBase Review
- Kam Taghizadeh Week 9
- Kam Taghizadeh Week 10
- Kam Taghizadeh Week 11
- Kam Taghizadeh Week 12
- Kam Taghizadeh Week 14