Owen R. Dailey Week 5
Template
User Page
Assignments
Individual Journals
The D614G Research Group Week 12
The D614G Research Group Week 14
Class Journals
Purpose
The purpose of this lab is to analyze the structure of the spike protein, the structure of the ACE2 protein, and the residues that allow them to interact/bind to one another. Additionally, the purpose is to view the different levels of the protein structure (primary, secondary, and tertiary structure).
Methods/Results
Exploring the Spike protein Structure
Exploring UniProt
- To begin, I navigated to UniProt Knowledgebase (UniProt KB), and I searched for the SARS-CoV-2 spike protein using the keyword "SARS-CoV-2" in the main UniProt search field
- This search yielded 1,616 search results; however, only 18 of these results were curated in Swiss-Prot
- Additionally, the vast majority of the results were viral proteins (1,611results), but four results were human proteins and one result was a mouse protein
- I then analyzed the search result with accession number "P0DTC2" which corresponded to the reference entry for the SARS-CoV-2 spike protein.
- Included information on the SARS-COV-2 spike protein:
- Function of the protein
- Names and taxonomy
- Subcellular location
- Pathology and biotech
- Posttranslational modification and processing
- Interactions
- Structure
- Family and domains
- Sequence
- Similar proteins
- Cross-references
Analyzing SARS-CoV RBD (optimized for human ACE2 recognition) and human ACE2: 3SCI
- I then viewed one of the structures of the SARS-CoV spike protein from Wan et al. (2020) in the NCBI Structure Database using the web-based iCn3D viewer
- The structure I chose to view was Figure 2C SARS-CoV RBD (optimized for human ACE2 recognition) and human ACE2: 3SCI
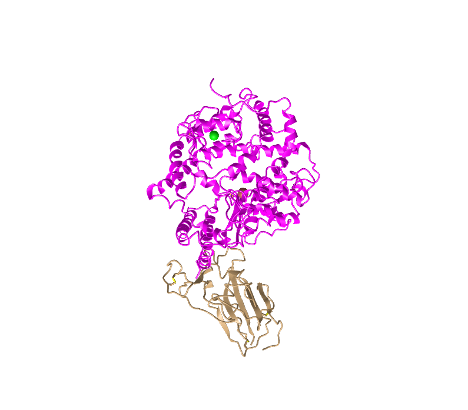
Standard Style
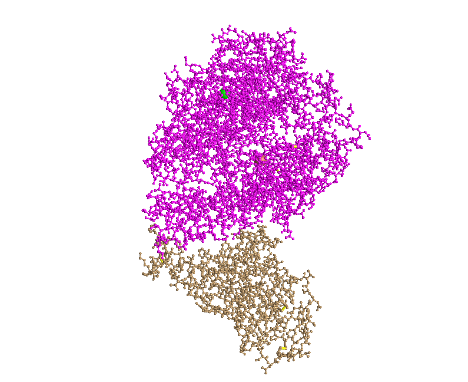
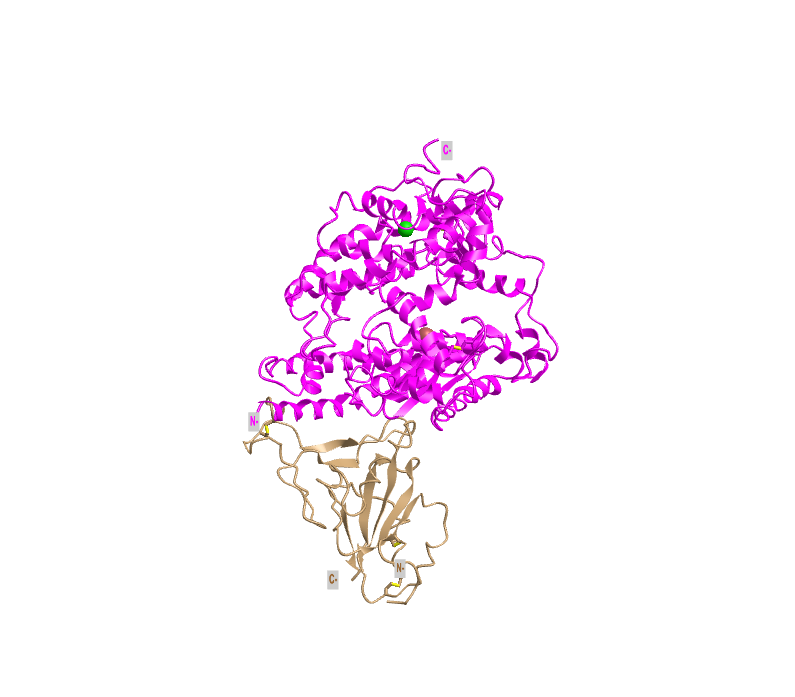
- I created a view of the protein that recreated Figure 1A from the Wan et al. (2020) paper by rotating the structure:
- This style shows the ACE2 protein in pink and the spike protein in tan (this will be the case for the following styles until the sphere styler)
- This is a tertiary structure because it shows the protein structure in three-dimensional space and does not just include the secondary structures
- There are two domains in this image because each protein (S protein and ACE2 protein) is one domain (scissor and hula-hoop test)
- There is not a place where you can cut either protein and have an independently folding domain
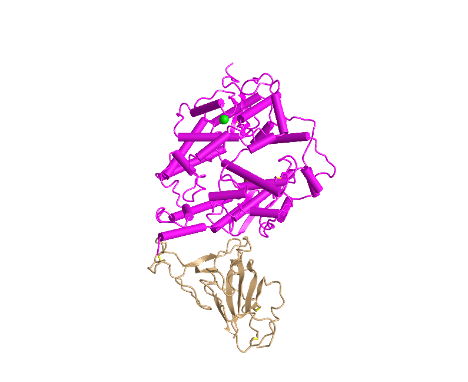
Cylinder and Plate Style
- I clicked on the Style > Proteins menu and selected Cylinder and Plate
- This style depicts the alpha-helices as cylinders and beta-sheets as plates
- This style is unique because it makes it very easy to see the secondary structures of each protein
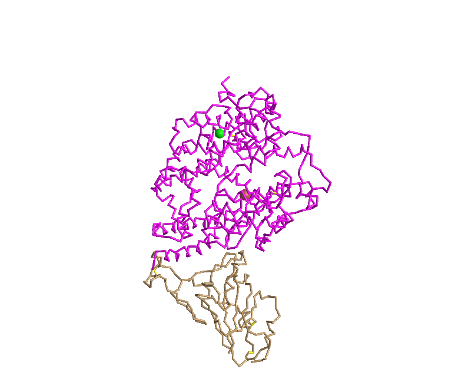
C Alpha Trace
- I clicked on the Style > Proteins menu and selected C Alpha Trace
- This style is unique because it shows the structure of the protein backbone because each "jag" represents the alpha Carbon in the protein backbone
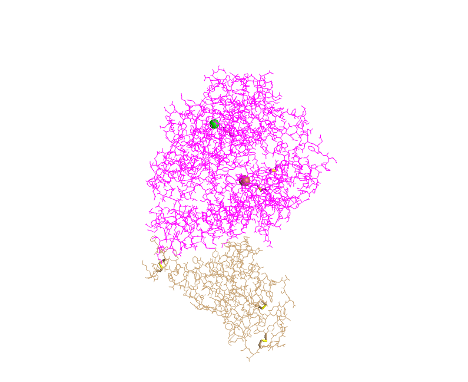
Lines
- I clicked on the Style > Proteins menu and selected Lines
- This style is unique because it shows the line structure of the protein backbone and the side chains that are protruding from it
- This style allows me to see the orientation of the side chains
Ball and Stick
- I clicked on the Style > Proteins menu and selected Ball and Stick
- This style is unique because it shows the protein backbone and its residues as the molecular ball and stick model
- This style allows one to see the 3D model of the atoms and their bonds that make up the protein
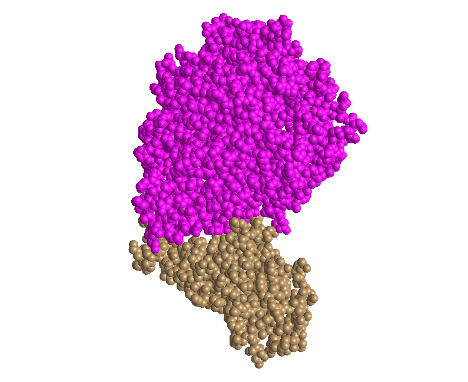
Spheres
- I clicked on the Style > Proteins menu and selected Spheres
- This style is unique because it depicts each amino acid as a sphere
- This style allows for easy visualization of the 3D space taken up by each amino acid
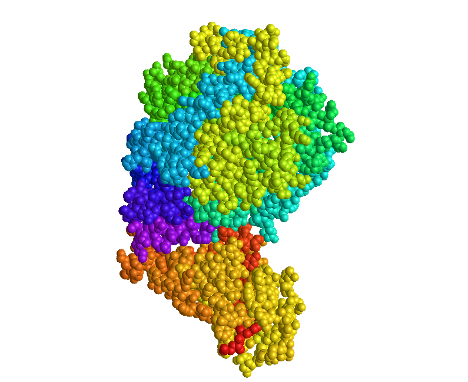
Spheres Spectrum Color
- In the “Spheres” view, I clicked on the Color menu and selected the Spectrum color scheme
- This color scheme is unique because it shows the ACE2 protein in a color spectrum from purple/blue (n-terminus) to green (c-terminus) and it shows the Spike protein in a color spectrum from yellow (n-terminus) to red (c-terminus)
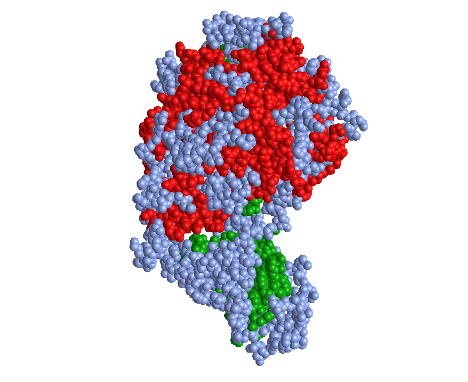
Spheres Secondary Color
- In the “Spheres” view, I clicked on the Color menu and selected the Secondary color scheme
- This color scheme is unique because it shows the amino acids involved in alpha helixes in the color red and the amino acids involved in beta sheets in green
- This sty;e allows one to view the amino acids involved in secondary structures with ease
Spheres Charge Color
- In the “Spheres” view, I clicked on the Color menu and selected the Charge color scheme
- This color scheme is unique because it shows the positively charged amino acids in blue, negatively charged amino acids in red, and neutrally charged amino acids in grey
Spheres Atom Color
- In the “Spheres” view, I clicked on the Color menu and selected the Atom color scheme
- This style is unique because it depicts carbons as grey spheres, oxygens as red spheres, nitrogens as blue spheres, and sulfurs as yellow spheres
General Questions/Answers
N-Terminus and C-Terminus
- The N-terminus of the spike protein and ACE2 protein was located
- In this image, the N-termini and C-termini are denoted by grey boxes on the spike protein and ACE2 protein
Secondary Structures
- Alpha helices are found in the ACE2 protein
- This was determined by observing the cylinder and plate style structure
- There were only cylinders
- Beta sheets are found in the spike protein
- This was determined by observing the cylinder and plate style structure
- There were only plates
Exploring the Civet ACE2-Spike Protein Structure
- First, I navigated to this website
- I then clicked on the Windows menu to “View Sequences & Annotations”
- In the window that appeared on the right, I clicked on the “Details” tab to show the actual amino acid sequences
- There were 2 sets of ACE2-spike proteins because of the way the proteins crystallized
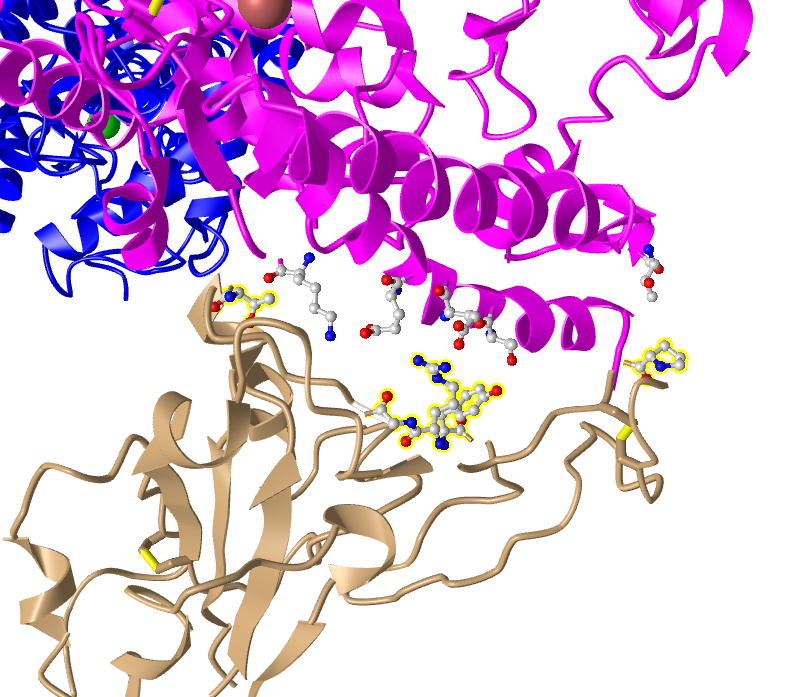
- I focused on the pink and tan chains and oriented them as they were shown in Figure 4B
- I followed these steps to make the amino acid side chains shown in the figure visible
- In the sequence window I went to sequence “Protein 3SCK_A” (in pink) and selected the following amino acids
- T31
- E35
- E38
- T82
- K353
- The part of the ribbon that represented these amino acids were highlighted in yellow in the structure
- I went to the Styles menu and selected Proteins > Ball and Stick
- I went to the Color menu and select Atom
- In the sequence window I went to sequence “Protein 3SCK_A” (in pink) and selected the following amino acids
- I repeated this process for the tan spike protein sequence 3SCK_E for the following amino acids:
- T487
- R479
- G480
- Y442
- P472
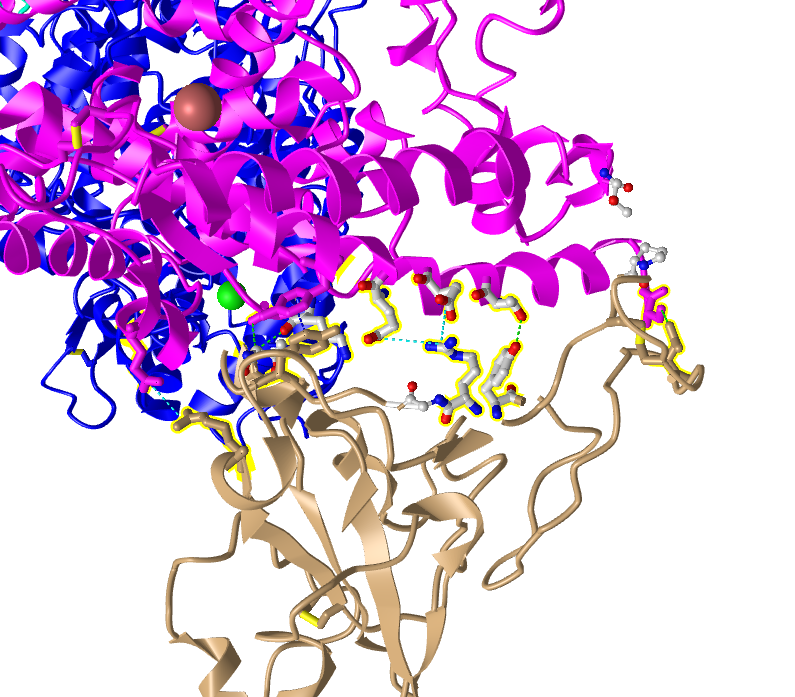
- E35 on ACE2 makes an ionic bond with R479 on spike protein
- E35 is acidic because it has a lot of red (oxgens) in the atom
- R479 is basic because it has a lot of blue (nitrogen) in the atom
- T31 on ACE2 makes a hydrogen bond with Y442 on spike protein
- T is polar because it has oxygens
- Y is hydrophobic because it has nitrogens
- I then went to the View menu and selected H bonds & Interactions
- In part 1 of the window that appeared, I unchecked “Contacts/Interactions” leaving Hydrogen Bonds and Ionic Interaction checked
- In part 2 of the window, I selected the first set “3SCK_A” (pink)
- In part 3 of the window, I selected the second set “3SCK_E” (tan)
- In part 4 of the window, I clicked the button “3D Display”
- This inserted dashed lines representing the ionic bonds and H-bonds between the two polypeptide chains and amino acids that were described above
Beginning your research project
- What question will you answer about sequence-->structure-->function relationships in the spike and/or ACE2 protein?
- To what extent are mutations in the human spike protein related to geography and time?
- What sequences will you use?
- China: NC_045512 (collected December 2019)
- Spain: MT956913 (collected April 15th)
- Italy: MT_483879 (collected March 18th)
- Washington, USA: MT598633 (collected February 20th)
- India: MT940464 (collected July 27th)
- New Zealand: MT706050 (collected March 21st)
- China: MT911467 (collected August 14th)
- Peru: MW030279 (collected May 6th)
Conclusion
By analyzing the structure of the spike protein, the structure of the ACE2 protein, and the residues that allow them to interact/bind to one another, I was able to conclude that the spike protein is relatively small compared to the ACE2 protein. Furthermore, I concluded that a relatively small number of amino acids in the spike protein and ACE2 protein interact with one another. Finally, I was able to conclude that the ACE2 protein contains a large number of alpha-helices, and the spike protein contains a large number of beta-sheets.
Aknowledgements
- I contacted/discussed with my homework partner, Ian Wright, via text and zoom to discuss the our research project and the domains of the spike and ACE2 protein
- I talked with my professor, Dr. Dahlquist, over zoom regarding the upcoming research project
- I copied and modified the procedures shown on the Week 5 Page
- I used the Wan et al. (2020) paper compare my protein structures
- I used sequences from GenBank
- I used NCBI Structure Database to view the protein structures
- Except for what is noted above, this individual journal entry was completed by me and not copied from another source
Owen R. Dailey (talk) 16:51, 7 October 2020 (PDT)
References
- Foldit - Blogs. (2020). Retrieved October 7, 2020, from https://fold.it/portal/blog
- Foldit - Solve Puzzles for Science. (2020). Retrieved October 7, 2020, from https://fold.it/portal/
- iCn3D: Web-based 3D Structure Viewer 3SCI. (2020). Retrieved October 1, 2020, from https://www.ncbi.nlm.nih.gov/Structure/icn3d/full.html?&mmdbid=97063&bu=1&showanno=1&source=full-feature
- iCn3D: Web-based 3D Structure Viewer 3SCK. (2020). Retrieved October 6, 2020, from https://www.ncbi.nlm.nih.gov/Structure/icn3d/full.html?pdbid=%203SCK
- OpenWetWare. (2020). BIOL368/F20:Week 5. Retrieved October 1, 2020, from https://openwetware.org/wiki/BIOL368/F20:Week_5
- Uniprot. (2020). S - Spike glycoprotein precursor - Severe acute respiratory syndrome coronavirus 2 (2019-nCoV) - S gene & protein. Retrieved October 1, 2020, from https://www.uniprot.org/uniprot/P0DTC2
- Wan, Y., Shang, J., Graham, R., Baric, R., & Li, F. (2020). Receptor Recognition by the Novel Coronavirus from Wuhan: an Analysis Based on Decade-Long Structural Studies of SARS Coronavirus. Journal Of Virology, 94(7). doi: 10.1128/jvi.00127-20