IGEM:Harvard/2006/Cyanobacteria
<html><style type='text/css'> .tabs {
font-size:80%;
font-weight:none;
width: 100%;
color: #FFFFFF;
background:#FFFFFF url("/images/5/54/DarkgreenTab-bg.gif") repeat-x bottom;
}
.tabs li {
background:url("/images/3/36/DarkgeenTab-left.gif") no-repeat left top;
}
.tabs a,.tabs strong {
background:url("/images/d/d3/DarkgreenTab-right.gif") no-repeat right top;
color:#FFFFFF;
padding: 3px 10px 3px 4px;
}
.tabs strong{
color:#CCFF00;
background-image:url("/images/b/b1/DarkgreenTab-right_on.gif");
}
.tabs a:hover{
color:#66FF00;
}
</style></html>
Introduction
Welcome to the lab notebook for the Cyanobacteria oscillator project! The goal of our team, composed of four members, is to reconstruct the cyanobacterial circadian oscillator system into E. coli. Three proteins, KaiA, B, and C, have been shown to have an in-vitro phosphorylation state oscillation (Nakajima et al. 2005) by transcriptional-translational independent methods. If this system can be reconstituted in E. coli, there are two important applications:
- Synthetic Biology: Creating a functional, oscillating set of proteins is the next logical step from the synthetic "repressilator" system engineered by Elowitz et al. (2000). Although a good proof of concept, the "repressilator" lacks the stability needed from a robust oscillator such as the naturally evolved cyanobacterial oscillator. This robust oscillator could prove useful in an eventual biocircuit.
- Circadian Biology: Cyanobacteria are the simplest model organisms for the study of circadian oscillation. Although circadian oscillation has been fairly well characterized, less is understood at the molecular level. By porting the oscillation system into E. coli, one can begin to understand more precisely the pathways involved in the genomic oscillation of cyanobacteria.
For more background information on the ciracadian system, please check out our "Literature" section. Otherwise, day-to-day work can be found under the "Lab Notebook" tab; we will post major results of our work and links to the days as they become available. If you have questions or comments, feel free to contact us: information is located at the main Harvard iGEM 2006 page. Thanks!
Sincerely,
Zhipeng, Hetmann, Dave, and Jeff
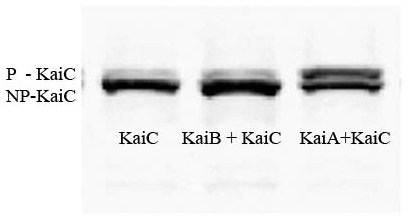
Update 10/27/06: We believe we can express the three proteins into e. coli, and that there is interaction between A+C and possible interaction between B+C. See the Lab Notebook for more information.
Outline of Findings and Signifigant Dates
- 07/05/06: The incubator for growing up our cyanobacteria is complete; we have cultures growing! Link
- 07/10/06: Some computer modeling has been done to see the effect of multiple unsyncronized clocks on phosphorylation state output. Link
- 07/21/06: Upon having trouble with site-specific mutagenesis on the KaiA and KaiBC operons from the cyanobacterial genome, we have decided to pursue synthesis of the constructs in parallel with continued extraction attempts. Link
- 08/01/06: Preliminary success with site-specific mutagenesis. Link
- 08/05/06: Promoter leakness tests come out negative. May have to use low-copy plasmids if we want good control of protein expression in Top10F. Link
- 08/11/06: We are moving to the synthetic KaiA, KaiB, and KaiC for future work. Link
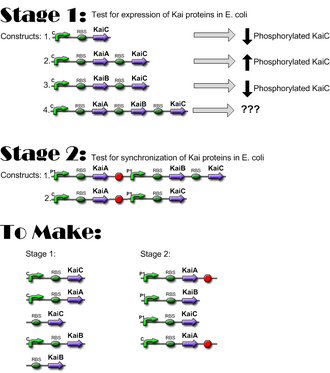
- 08/30/06: We successfully made the first construct, Lac+RBS+KaiC. Link
- 09/01/06: Using the newly developed ligation protocol, we have successfully repeated Lac+RBS+KaiC from 08/30/06 and made Lac+RBS+KaiA. Link
- 10/21/06: Successfully made Lac+RBS+KaiB and Lac+RBS+KaiA+Lac+RBS+KaiC. Link
- 10/24/06: Successfully made Lac+RBS+KaiB+Lac+RBS+KaiC. Link
- 10/25/06: Constructs for Stage I have been completed; ready to move to Stage I of Western Blotting, to verify expression of KaiC and interaction of KaiA and KaiB with KaiC. Link
- 10/27/06: Preliminary data indicates that the Kai proteins are being expressed in e. coli and that there is interaction between the three proteins! Link
Construct Planning
Lengths
From VF2 to VR (BioBrick primers):
- KaiA + J04500: 1406 bp
- KaiB + J04500: 859 bp
- KaiC + J04500: 2110 bp
Agenda
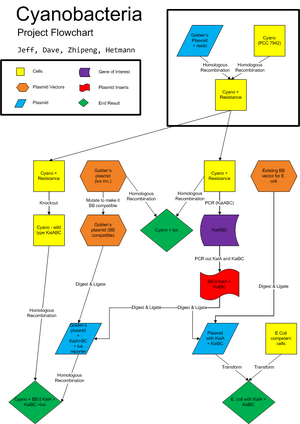
See image at right for our long-term project outline.
BioBricks Used
- <bbpart>BBa_J04450</bbpart>
- RFP device
- Insert size: 1069bp
- [pSB1A2]
- High-copy, AmpR
- Size: 2079bp
- <bbpart>BBa_J04500</bbpart>
- Lac promoter + RBS
- Insert size: 220bp
- [pSB1AK3]
- High-copy, AmpR, KanR
- Insert size: 3189bp
- [pSB4A3]
- Low-copy, AmpR
- Insert size: 3339 bp
- <bbpart>BBa_R0010</bbpart> + <bbpart>BBa_E0241</bbpart>
- GFP device
- Insert size: 995 bp
- <bbpart>BBa_J04450</bbpart>
Presentations
- Project proposal (week 2)
- Week 3 progress update
- Built incubator and obtained WH8102, PCC7942, and PCC6803 strains
- Week 4 progress update
- Week 5 progress update, upd. 10:10 7/17
- Week 6 progress update, upd. 10:02 7/24 HH
- Week 7 progress update
- Week 8 progress update
- Week 9 progress update
- Week 10 progress update, 50% complete
- Final Presentation (incomplete) --old
- Final Presentation (complete) --old
- Jamboree presentation (in progress)
- Script (in progress)
- Cyano poster (in progress)
Team Members
- Hetmann Hsieh (talk, edits)
- Jeffrey Lau (talk, edits)
- Zhipeng Sun (talk, edits)
- David Ramos (talk, edits)
Recent Changes
List of abbreviations:
- N
- This edit created a new page (also see list of new pages)
- m
- This is a minor edit
- b
- This edit was performed by a bot
- (±123)
- The page size changed by this number of bytes