IGEM:Harvard/2006/Cyanobacteria/Bioinformatics
<html><style type='text/css'> .tabs {
font-size:80%;
font-weight:none;
width: 100%;
color: #FFFFFF;
background:#FFFFFF url("/images/5/54/DarkgreenTab-bg.gif") repeat-x bottom;
}
.tabs li {
background:url("/images/3/36/DarkgeenTab-left.gif") no-repeat left top;
}
.tabs a,.tabs strong {
background:url("/images/d/d3/DarkgreenTab-right.gif") no-repeat right top;
color:#FFFFFF;
padding: 3px 10px 3px 4px;
}
.tabs strong{
color:#CCFF00;
background-image:url("/images/b/b1/DarkgreenTab-right_on.gif");
}
.tabs a:hover{
color:#66FF00;
}
</style></html>
Synthesis
IGEM:Harvard/2006/Cyanobacteria_synth
For synthesis, we are probably going to go with GeneART. I created three constructs for each protein. They are the coding sequence + 22 bp biobricks prefix + 21 bp biobricks suffix + extra stop codon. Also, point mutagenesis on two sites was conducted on the kaiC construct to match those in experiments. For KaiA, the GTG start was kept.
For each construct, useful information:
- KaiA
- sequence in rtf format. Click here.
- pdf preview #1. Click here.
- pdf preview #2. Click here.
- .gb file. Click here.
- Proof of allignment using AllignX. Click here.
- KaiB
- sequence in rtf format. Click here.
- pdf preview #1. Click here.
- pdf preview #2. Click here.
- .gb file. Click here.
- Proof of allignment using AllignX. Click here.
- KaiC
- sequence in rtf format. Click here.
- pdf preview #1. Click here.
- pdf preview #2. Click here.
- .gb file. Click here.
- Proof of allignment using AllignX. Click here.
Useful Files
KABC stuff
The other FASTA/Vector NTI 7/ random files:
- The PCC7942 genome downloaded from NCBI in FASTA format. (Very Large!) Click here.
- The KaiA, KaiB, and KaiC sequences downloaded from NCBI. In FASTA format but combined. Click here.
- The Vector NTI file with the PCC7942 genome, along with annotations by Peng of the location of KaiABC/promoter/primers/restriction sites/etc... Use this if you are going to be adding information to our sequence! Click here.
- The Vector NTI file with the KaiABC area of interest extracted. Not really necessary but easier to see then the Vector NTI file. Click here.
- Scrap file by Peng when designing primers. Click here.
- Link to Kun's Oligonucleotide calculator. Click here.
- Link to DNA sequence inverser. Click here.
Topo stuff (7/17)
- The PCR-bluntII Topo vector sequence was found on the invitrogen website under support-->vector info.
- The topo vector + our insert in .gb format. Click here.
- The pdf of the topo vector + our insert, w/ restriction sites Click here.
Primer Design
Primer Set #1: Extraction of kABC and Site Mutagenesis (10)
Misc. Info
According to BLAST, KaiC (519aa) is only 81% identical between PCC7942 and 6803. With the info that PCC7942 is the main working strand, I propose that we only work with PCC7942 for now and if we have time do research into 6803 - it would need a whole new set of primers.
Basically, the information on this page is for designing the primers used to Colony PCR out the kaiABC construct from the PCC7942 strain, and the primers used for site-directed mutagenesis and Crossover PCR. The site-directed mutagenesis is used to remove the 1 EcoRI sites and the 2 PstI sites that interfere with BioBrick construction.
The task is less than trival however. There are 3 nearby PstI sites, and two of them are right by the kaiA promoter region - one is -25 to -30 away and one is -76 to -82 away. This means that whereas we can mutate the -25 to -30 region, we don't know if it will affect sigma binding specificity. Additionally, the -76 to -82 will have to be excluded while allowing for a region big enough to be seen on a gel after crossover PCR. I resolve this by using two seperate primers at the end; a extraction forward/reverse and a crossover forward/reverse.
Background Papers
The background papers mainly deal with identifying the promoter sequences for KaiBC (one operon) and KaiA (one operon). Ultimately, it looks as if they fall under the same -35/-10 region as E. coli; it was identified that -55 to 14 is important for KaiBC, which I will assume holds true for KaiA in primer design.
These papers might have our promoter sequences:
- Ishiura M, Kutsuna S, Aoki S, Iwasaki H, Andersson CR, Tanabe A, Golden SS, Johnson CH, and Kondo T. Expression of a gene cluster kaiABC as a circadian feedback process in cyanobacteria. Science. 1998 Sep 4;281(5382):1519-23. DOI:10.1126/science.281.5382.1519 |
- Kutsuna S, Nakahira Y, Katayama M, Ishiura M, and Kondo T. Transcriptional regulation of the circadian clock operon kaiBC by upstream regions in cyanobacteria. Mol Microbiol. 2005 Sep;57(5):1474-84. DOI:10.1111/j.1365-2958.2005.04781.x |
- Goto-Seki A, Shirokane M, Masuda S, Tanaka K, and Takahashi H. Specificity crosstalk among group 1 and group 2 sigma factors in the cyanobacterium Synechococcus sp. PCC7942: In vitro specificity and a phylogenetic analysis. Mol Microbiol. 1999 Nov;34(3):473-84. DOI:10.1046/j.1365-2958.1999.01608.x |
- Especially paper #3, which states that there are 4 known sigma factors.
- Paper #2 says that -55 to 14 was experimentally determined to be the promoter/operon area for kaiBC (not kaiA :(); -35 and -10 have homology to sigma 70 in E. coli
Proposed Primers
Below are 10 proposed primers:
# 7942_KABC_extF
# 7942_KABC_extR
# 7942_KABC_crossF
# 7942_KABC_crossR
# 7942_KABC_pst1R
# 7942_KABC_pst1F
# 7942_KABC_pst2R
# 7942_KABC_pst2F
# 7942_KABC_eco1R
# 7942_KABC_eco1F
First, a .jpg of the primers/procedure. Also available as a .ppt here
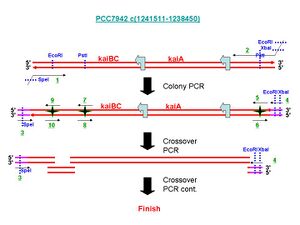
# 7942_KABC_extF (1)
This primer is used for the first Colony PCR. In the Picture, it is noted as an underlined "1" in green.
The design is a hybrid primer. with a part with:
- Homology with PCC7942 in location(1238522-1238546)
- Same with top strand of PCC7942
- Biobricks homology, with S site and N site.
- Homology with PCC7942 in location(1238522-1238546)
1)
5’ GACAAGAATACCTAAGCGCGATCGC 3’ (FORWARD) LENGTH=25BP TM = 61.1C GC =52.0% LOCATION(1238522-1238546) By end of KaiC
2)
For area by end of KaiC: 5’ AGCGGCCGCTACTAGTAA 3’ (contains SpeI and NotI + AG)
Then, final primer is:
5’ AGCGGCCGCTACTAGTAA GACAAGAATACCTAAGCGCGATCGC ‘3 (Tm=77.67)
Checked for repetition in genome.
# 7942_KABC_extR (2)
This primer is used for the first Colony PCR. In the Picture, it is noted as an underlined "2" in green. The design is a hybrid primer. with a part with:
- Homology with PCC7942 in locationc(1241538-1241511), where c = complement
- Same with bottom strand of PCC7942
- Biobricks homology, with E site and N site and X site.
- Homology with PCC7942 in locationc(1241538-1241511), where c = complement
1)
5’ AGTGCTAGGCTAAATTAAATTTTTCC 3’ Length = 26bp Locationc(1241489-1241464) Tm = 60.13 GC 30.8% By KaiA promoter Contains a PstI site
2)
For area by KaiA promoter: 5’ GAATTCGCGGCCGCTTCTAGAGT 3’ (contains EcoRI + NotI + XbaI + ‘GT’)
Then, final primer is:
5’ GAATTCGCGGCCGCTTCTAGAGT AGTGCTAGGCTAAATTAAATTTTTCC ‘3 (Tm = 74.54)
Checked for repetition in genome.
# 7942_KABC_crossF (3)
This primer is based off of #7942_KABC_extF (1), but does not have the first area of homology to PCC7942. This is made such that it has the same approximate melting temperature as #7942_KABC_crossR (4). It is a green underlined (3) in the picture.
The primer is:
5’ AGCGGCCGCTACTAGTAA 3’ Tm = 60.87
Checked for repetition in genome.
# 7942_KABC_crossR (4)
Prehaps the trickest primer to understand, this primer is here to be used in the Crossover PCR instead of #7942_KABC_extR; normally, one would use the latter but we have to use the former due to 2 PstI sites close to what we want to amplify. Artificially, we can then avoid incorporating one PstI site, and mutate the other while allowing for a segment large enough to be seen during gel purification.
It is a green underlined (4) in the picture.
The primer is:
5’ GAATTCGCGGCCGCTTC 3’ Tm = 62.75
Checked for repetition in genome.
# 7942_KABC_pst1R (5)
This primer is to mutate the PstI site at 1243438 (eg. -27 away from KaiA). Hopefully this will not interfere with the sigma binding specificity. It is a mutation in a non-coding site. Turns a T into a G on the dominant strand.
Primer is:
5’ tcaggactgagtcGgcaga 3’ Tm = 62.96
Checked for repetition in genome. Location(1241425-1241443)
# 7942_KABC_pst1F (6)
Complement to #7942_KABC_pst1R (5).
Primer is:
5’ TCTGCCGACTCAGTCCTGA 3’ Tm = 62.96
Checked for repetition in genome.
# 7942_KABC_pst2R (7)
Thi=s primer is to mutate the PstI site at 1239051 (in the KaiC coding region). Silent mutation of an A to a G. Keeps amino acid Serine.
Primer is:
5’ caagacctgcGgattcaggata 3’ Tm = 62.57
Checked for repetition in genome. Location(1239041-1239062)
# 7942_KABC_pst2F (8)
Complement to #7942_KABC_pst2R (7).
Primer is:
5’ TATCCTGAATCCGCAGGTCTTG 3’ Tm = 62.57
Checked for repetition in genome.
# 7942_KABC_eco1R (9)
This primer is to mutate the EcoRI site at 1238703 (in the KaiC coding region). Silent mutation of an T to a C. Keeps amino acid Glu.
Primer is:
5’ ctgatcatgaaCtcgcggattg 3’ Tm = 62.36
Checked for repetition in genome. Location (1238692-1238713)
# 7942_KABC_eco1F (10)
Complement to #7942_KABC_eco1R (9).
Primer is:
5’ CAATCCGCGAGTTCATGATCAG 3’ Tm = 62.36
Checked for repetition in genome.
Final Primer Summary
If the top primers are okay, the following will be sent to Alain for ordering on 6/30:
GAATTCGCGGCCGCTTCTAGAGTAGTGCTAGGCTAAATTAAATTTTTCC GAATTCGCGGCCGCTTC AGCGGCCGCTACTAGTAAGACAAGAATACCTAAGCGCGATCGC AGCGGCCGCTACTAGTAA ctgatcatgaaCtcgcggattg CAATCCGCGAGTTCATGATCAG caagacctgcGgattcaggata TATCCTGAATCCGCAGGTCTTG tcaggactgagtcGgcaga TCTGCCGACTCAGTCCTGA
Primer Set #2: Sequencing KABC
This set of primer design is for sequencing the extract we obtained on 7/10 of kABC; to do so we need primers which will chop the sequence up into ~500bp segments. See Primer Set #1 for the sequence we are working with.
Because kABC is on the complement strand, I propose that it will be easier for us to work with sequencing on the direct strand. eg we will design primers to be the complement of the coding strand.
1238522 --a1--- 1239000 ---a2--- 1239501 --a3--- 1240002--a4- 1240503 -a5--- 1241001 ---a6-1241469
(approx. #'s)
NOTE: Edit 7/13: changed A4
Primers we will need to order
kabc_seqA1: 5' ATTGCAATACGAGCTGGCTT 3'
- location(1238980-1238999), tm 51.8C
kabc_seqA2: 5' ACGTTGCGGAGAATCACGAC 3'
- location (1239481-1239500), tm 54.9C
kabc_seqA3: 5' ACCACTAACGAGGGTCGATC 3'
- location (1239972-1239991), tm 50.5C
kabc_seqA4: 5' GGGGTAGCAACAGCAATCAA 3'
- location(1240480-1240499), tm 52.6C
kabc_seqA5: 5' GCCCCCATCAGCATGATGTG 3'
- location(1240984-1241003), tm 58.2C
Primers we have
For the forward sequence, the biobricks forward primer (or the topo primer).
Primers we should order anyways (reverse complement seqs)
kabc_seqA1_RC: 5' AAGCCAGCTCGTATTGCAAT 3'
kabc_seqA2_RC: 5' GTCGTGATTCTCCGCAACGT 3'
kabc_seqA3_RC: 5' GATCGACCCTCGTTAGTGGT 3'
kabc_seqA4: 5' TTGATTGCTGTTGCTACCCC 3'
kabc_seqA5_RC: 5' CACATCATGCTGATGGGGGC 3'
Final Primer Order
kabc_seqA1: 5' ATTGCAATACGAGCTGGCTT 3'
kabc_seqA2: 5' ACGTTGCGGAGAATCACGAC 3'
kabc_seqA3: 5' ACCACTAACGAGGGTCGATC 3'
kabc_seqA4: 5' GGGGTAGCAACAGCAATCAA 3'
kabc_seqA5: 5' GCCCCCATCAGCATGATGTG 3'
kabc_seqA1_RC: 5' AAGCCAGCTCGTATTGCAAT 3'
kabc_seqA2_RC: 5' GTCGTGATTCTCCGCAACGT 3'
kabc_seqA3_RC: 5' GATCGACCCTCGTTAGTGGT 3'
kabc_seqA4_RC: 5' TTGATTGCTGTTGCTACCCC 3'
kabc_seqA5_RC: 5' CACATCATGCTGATGGGGGC 3'
Primer Set #3: Extracting KaiA,BC
These primers will be made to extract the kaiABC coding sequences w/ biobricks homology attached.
KaiA coding
This primer is designed to extract the KaiA coding sequence from the start codon until the end of the sequence. The ends have biobricks homology. Terminator is NOT included; in cyanobacteria it looks to be rho-dependent.
Targeted location: comp(1241411-1240557)
F-primer: kabc_extractA_F
Area with homology to kaiA start: 5' gtgctctcgcaaattgcaatctgc 3' comp(1241411-1241388) length:26 Tm: 61.8 GC%: 50
Area with homology to biobricks: 5' GAATTCGCGGCCGCTTCTAGAGT 3' (contains EcoRI + NotI + XbaI + ‘GT’)
Final kabc_extractA_F: 5' GAATTCGCGGCCGCTTCTAGAGTgtgctctcgcaaattgcaatctgc 3' length=47 Tm: 83.0
R-primer: kabc_extractA_R
Area with homology to kaiA end: 5' TCAGGTTTCTCGTGGGATAGACCGT 3' location(1240557-1240581) length=25 Tm: 60.9 GC%: 52
Area with homology to biobricks 5’ AGCGGCCGCTACTAGTAA 3’ (contains SpeI and NotI + AG)
Final kabc_extractA_R: 5' AGCGGCCGCTACTAGTAATCAGGTTTCTCGTGGGATAGACCGT 3' length=43 Tm: 76.7
KaiBC coding + non-coding in the middle
This primer is designed to extract the KaiBC coding sequence from the start of kaiB until the end of KaiC. The ends have biobricks homology. Terminator is NOT included; in cyanobacteria it looks to be rho-dependent. Non-coding insert is included.
Targeted location: comp(1240467-1238550)
F-primer: kabc_extractBC_F
Area with homology to kaiB start: 5' atgagccctcgtaaaacctacattctca 3' comp(1240467-1240440) length:28 Tm: 60.4 GC%: 42.3
Area with homology to biobricks: 5' GAATTCGCGGCCGCTTCTAGAGT 3' (contains EcoRI + NotI + XbaI + ‘GT’)
Final kabc_extractBC_F: 5' GAATTCGCGGCCGCTTCTAGAGTatgagccctcgtaaaacctacattctca 3' length=49 Tm: 79.6
R-primer: kabc_extractBC_R
Area with homology to kaiC end: 5' ctagctctccggccctttttcttga 3' location(1238550-1238574) length=25 Tm: 61.9 GC%: 52
Area with homology to biobricks 5’ AGCGGCCGCTACTAGTAA 3’ (contains SpeI and NotI + AG)
Final kabc_extractBC_R: 5' AGCGGCCGCTACTAGTAActagctctccggccctttttcttga 3' length=43 Tm: 77
KaiB coding
This primer is designed to extract the KaiB coding sequence from the start of kaiB until the end of kaiB. The ends have biobricks homology. Terminator is NOT included; in cyanobacteria it looks to be rho-dependent. NOTE: ONLY KAIB!
Targeted location: comp(1240467-1240159)
F-primer: Use kabc_extractBC_F
R-primer: kabc_extractB_only_R
Area with homology to kaiB end: 5' ttagaagtcgtcggaatcttgaagttcg 3' location(1240159-1240186) length=28 Tm: 61.0 GC%: 42.9
Area with homology to biobricks 5’ AGCGGCCGCTACTAGTAA 3’ (contains SpeI and NotI + AG)
Final kabc_extractB_only_R: 5' AGCGGCCGCTACTAGTAAttagaagtcgtcggaatcttgaagttcg 3' length=46 Tm: 75.8
KaiC coding
This primer is designed to extract the KaiC coding sequence from the start of kaiC until the end of kaiC. The ends have biobricks homology. Terminator is NOT included; in cyanobacteria it looks to be rho-dependent. NOTE: ONLY KAIC!
Targeted location: comp(1240159-1238550)
R-primer: Use kabc_extractBC_R
F-primer: kabc_extractC_only_F
Area with homology to kaiC beginning: 5' atgacttccgctgagatgactagccc 3' c(1240109-1240084) length=26 Tm: 61.8 GC%: 53.8
Area with homology to biobricks: 5' GAATTCGCGGCCGCTTCTAGAGT 3' (contains EcoRI + NotI + XbaI + ‘GT’)
Final kabc_extractC_only_F: 5' GAATTCGCGGCCGCTTCTAGAGTatgacttccgctgagatgactagccc 3' length=49 Tm: 79.6
Final Primers to Order
kabc_extractA_F: 5' GAATTCGCGGCCGCTTCTAGAGTgtgctctcgcaaattgcaatctgc 3' kabc_extractA_R: 5' AGCGGCCGCTACTAGTAATCAGGTTTCTCGTGGGATAGACCGT 3' kabc_extractBC_F: 5' GAATTCGCGGCCGCTTCTAGAGTatgagccctcgtaaaacctacattctca 3' kabc_extractBC_R: 5' AGCGGCCGCTACTAGTAActagctctccggccctttttcttga 3' kabc_extractB_only_R: 5' AGCGGCCGCTACTAGTAAttagaagtcgtcggaatcttgaagttcg 3' kabc_extractC_only_F: 5' GAATTCGCGGCCGCTTCTAGAGTatgacttccgctgagatgactagccc 3'