IGEM:Harvard/2006/Cyanobacteria: Difference between revisions
From OpenWetWare
Jump to navigationJump to search
| Line 6: | Line 6: | ||
Welcome to the lab notebook for the Cyanobacteria project! The goal of our team, composed of four members, is to reconstruct the cyanobacterial circadian oscillator system into E. coli. Three proteins, KaiA, B, and C, have been shown to have an in-vitro phosphorylation state oscillation (Nakajima et al. 2005) by transcriptional-translational independent methods. If this system can be reconstituted in E. coli, there are two important applications: | Welcome to the lab notebook for the Cyanobacteria project! The goal of our team, composed of four members, is to reconstruct the cyanobacterial circadian oscillator system into E. coli. Three proteins, KaiA, B, and C, have been shown to have an in-vitro phosphorylation state oscillation (Nakajima et al. 2005) by transcriptional-translational independent methods. If this system can be reconstituted in E. coli, there are two important applications: | ||
#'''Synthetic Biology''': Creating a functional, oscillating set of proteins is the next logical step from the synthetic "repressilator" system engineered by Elowitz et al. (2000). Although a good proof of concept, the "repressilator" lacks the stability needed from a robust oscillator such as the naturally evolved cyanobacterial oscillator. This robust oscillator could prove useful in an eventual biocircuit. | |||
#'''Circadian Biology''': Cyanobacteria are the simplest model organisms for the study of circadian oscillation. Although circadian oscillation has been fairly well characterized, less is understood at the molecular level. By porting the oscillation system into E. coli, one can begin to understand more precisely the pathways involved in the genomic oscillation of cyanobacteria. | |||
==Construct Planning== | ==Construct Planning== | ||
Revision as of 11:58, 27 October 2006
<html><style type='text/css'> .tabs {
font-size:80%;
font-weight:none;
width: 100%;
color: #FFFFFF;
background:#FFFFFF url("/images/5/54/DarkgreenTab-bg.gif") repeat-x bottom;
}
.tabs li {
background:url("/images/3/36/DarkgeenTab-left.gif") no-repeat left top;
}
.tabs a,.tabs strong {
background:url("/images/d/d3/DarkgreenTab-right.gif") no-repeat right top;
color:#FFFFFF;
padding: 3px 10px 3px 4px;
}
.tabs strong{
color:#CCFF00;
background-image:url("/images/b/b1/DarkgreenTab-right_on.gif");
}
.tabs a:hover{
color:#66FF00;
}
</style></html>
Introduction
Welcome to the lab notebook for the Cyanobacteria project! The goal of our team, composed of four members, is to reconstruct the cyanobacterial circadian oscillator system into E. coli. Three proteins, KaiA, B, and C, have been shown to have an in-vitro phosphorylation state oscillation (Nakajima et al. 2005) by transcriptional-translational independent methods. If this system can be reconstituted in E. coli, there are two important applications:
- Synthetic Biology: Creating a functional, oscillating set of proteins is the next logical step from the synthetic "repressilator" system engineered by Elowitz et al. (2000). Although a good proof of concept, the "repressilator" lacks the stability needed from a robust oscillator such as the naturally evolved cyanobacterial oscillator. This robust oscillator could prove useful in an eventual biocircuit.
- Circadian Biology: Cyanobacteria are the simplest model organisms for the study of circadian oscillation. Although circadian oscillation has been fairly well characterized, less is understood at the molecular level. By porting the oscillation system into E. coli, one can begin to understand more precisely the pathways involved in the genomic oscillation of cyanobacteria.
Construct Planning
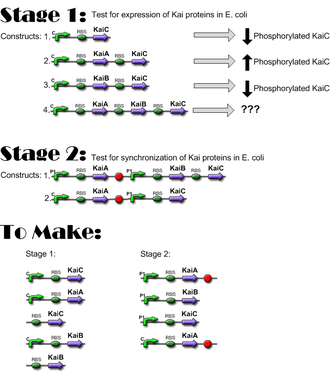
Lengths
From VF2 to VR (BioBrick primers):
- KaiA + J04500: 1406 bp
- KaiB + J04500: 859 bp
- KaiC + J04500: 2110 bp
Synthesis
Agenda
See image at right for our long-term project outline.

Click here for our current agenda
Kai Gene sizes
- KaiA size: 855bp
- 903bp with BioBrick ends
- KaiB: 309bp
- 357bp with BioBrick ends
- KaiC: 1560bp
- KaiA size: 855bp
BioBricks Used
- <bbpart>BBa_J04450</bbpart>
- RFP device
- Insert size: 1069bp
- [pSB1A2]
- High-copy, AmpR
- Size: 2079bp
- <bbpart>BBa_J04500</bbpart>
- Lac promoter + RBS
- Insert size: 220bp
- [pSB1AK3]
- High-copy, AmpR, KanR
- Insert size: 3189bp
- [pSB4A3]
- Low-copy, AmpR
- Insert size: 3339 bp
- <bbpart>BBa_R0010</bbpart> + <bbpart>BBa_E0241</bbpart>
- GFP device
- Insert size: 995 bp
- <bbpart>BBa_J04450</bbpart>
Presentations
- Project proposal (week 2)
- Week 3 progress update
- Built incubator and obtained WH8102, PCC7942, and PCC6803 strains
- Week 4 progress update
- Week 5 progress update, upd. 10:10 7/17
- Week 6 progress update, upd. 10:02 7/24 HH
- Week 7 progress update
- Week 8 progress update
- Week 9 progress update
- Week 10 progress update, 50% complete
- Final Presentation (incomplete) --old
- Final Presentation (complete) --old
- Jamboree presentation (in progress)
Team Members
- Hetmann Hsieh (talk, edits)
- Jeffrey Lau (talk, edits)
- Zhipeng Sun (talk, edits)
- David Ramos (talk, edits)
Recent Changes
List of abbreviations:
- N
- This edit created a new page (also see list of new pages)
- m
- This is a minor edit
- b
- This edit was performed by a bot
- (±123)
- The page size changed by this number of bytes
4 May 2024
| 18:10 | The Paper that Launched Microfluidics - Xi Ning diffhist −86 Xning098 talk contribs (→Electrokinetic effect) | ||||
| 11:36 | User:Jernej Turnsek diffhist +50 Jernej Turnsek talk contribs | ||||
|
|
09:20 | User:Massih Forootan 3 changes history +83 [Massih Forootan (3×)] | |||
|
|
09:20 (cur | prev) +78 Massih Forootan talk contribs (→Link to Publications) | ||||
|
|
09:18 (cur | prev) 0 Massih Forootan talk contribs (→Skills) | ||||
|
|
09:18 (cur | prev) +5 Massih Forootan talk contribs (→Skills) | ||||
|
|
07:28 | CHEM-ENG590E:Wiki Textbook 4 changes history −445 [Sarah L. Perry (4×)] | |||
|
|
07:28 (cur | prev) +84 Sarah L. Perry talk contribs (→Chapter 4 - Flow Control and Mixing) | ||||
|
|
07:21 (cur | prev) −490 Sarah L. Perry talk contribs (→Chapter 9 - Microfluidics and Cell Culture) | ||||
|
|
07:19 (cur | prev) −35 Sarah L. Perry talk contribs (→Chapter 9 - Microfluidics and Cell Culture) | ||||
|
|
07:18 (cur | prev) −4 Sarah L. Perry talk contribs (→Chapter 4 - Flow Control and Mixing) | ||||
| 07:18 | Move log Sarah L. Perry talk contribs moved page Microfluidic Gradient Generators - Greg Schneider to Microfluidic Gradient Generators (Adding an Author) | ||||
| 00:15 | Renhao Li Lab:Publications diffhist +265 Renhao Li talk contribs (→2024) | ||||
3 May 2024
| 15:02 | CHIP:Talks diffhist 0 Gabor Balazsi talk contribs | ||||
| 15:01 | CHIP:Data diffhist −23 Gabor Balazsi talk contribs | ||||
|
|
14:33 | UA Biophysics:Protocols:Kanamycin 5 changes history −421 [Elizabeth Suesca (5×)] | |||
|
|
14:33 (cur | prev) +13 Elizabeth Suesca talk contribs | ||||
|
|
14:21 (cur | prev) +16 Elizabeth Suesca talk contribs (→Materials) | ||||
|
|
14:19 (cur | prev) −6 Elizabeth Suesca talk contribs | ||||
|
|
14:14 (cur | prev) −50 Elizabeth Suesca talk contribs | ||||
|
|
14:13 (cur | prev) −394 Elizabeth Suesca talk contribs | ||||
|
|
14:33 | UA Biophysics:Protocols:Ampicillin 4 changes history −379 [Elizabeth Suesca (4×)] | |||
|
|
14:33 (cur | prev) +15 Elizabeth Suesca talk contribs | ||||
|
|
14:19 (cur | prev) −7 Elizabeth Suesca talk contribs | ||||
|
|
14:11 (cur | prev) 0 Elizabeth Suesca talk contribs (→PROTOCOL) | ||||
|
|
14:09 (cur | prev) −387 Elizabeth Suesca talk contribs | ||||
|
|
14:31 | UA Biophysics:Protocols:Erythromycin 3 changes history −315 [Elizabeth Suesca (3×)] | |||
|
|
14:31 (cur | prev) −7 Elizabeth Suesca talk contribs | ||||
|
|
14:30 (cur | prev) −12 Elizabeth Suesca talk contribs (→Materials) | ||||
|
|
14:18 (cur | prev) −296 Elizabeth Suesca talk contribs | ||||
|
|
14:29 | UA Biophysics:Protocols:Chloramphenicol 2 changes history −427 [Elizabeth Suesca (2×)] | |||
|
|
14:29 (cur | prev) +36 Elizabeth Suesca talk contribs | ||||
|
|
14:16 (cur | prev) −463 Elizabeth Suesca talk contribs | ||||
|
|
N 12:09 | BioMicroCenter:Oligo Synthesis 7 changes history +4,408 [Noelani Kamelamela (7×)] | |||
|
|
12:09 (cur | prev) −14 Noelani Kamelamela talk contribs (→Dr Oligo 96) | ||||
|
|
12:09 (cur | prev) −16 Noelani Kamelamela talk contribs (→STX-200) | ||||
|
|
12:08 (cur | prev) −46 Noelani Kamelamela talk contribs (→Dr Oligo 96) | ||||
|
|
12:08 (cur | prev) +35 Noelani Kamelamela talk contribs (→Dr Oligo 96) | ||||
|
|
08:29 (cur | prev) +7 Noelani Kamelamela talk contribs (→Dr Oligo 96) | ||||
|
|
08:27 (cur | prev) +59 Noelani Kamelamela talk contribs (→Dr Oligo 96) | ||||
| N |
|
08:16 (cur | prev) +4,383 Noelani Kamelamela talk contribs (Created page with "{{BioMicroCenter}} ''' ** All users must be trained before being allowed to use the equipment **'''<BR><BR> The BioMicro Center currently hosts two oligo synthesizers: one Dr Oligo 96 (Biolytic) and one Syntax STX-200 (DNAScript). <br><br> Dr Oligo is optimized for higher concentrations of oligos made through polyamidite synthesis. For ready to use oligos a series of steps must be completed using the other related machines in the Center including the column presser, the...") | |||
|
|
12:06 | BioMicroCenter:Covaris 5 changes history +172 [Noelani Kamelamela (5×)] | |||
|
|
12:06 (cur | prev) −5 Noelani Kamelamela talk contribs (→R230) | ||||
|
|
09:55 (cur | prev) +2 Noelani Kamelamela talk contribs (→R230) | ||||
|
|
09:54 (cur | prev) +1 Noelani Kamelamela talk contribs (→R230) | ||||
|
|
09:52 (cur | prev) +4 Noelani Kamelamela talk contribs (→R230) | ||||
|
|
08:41 (cur | prev) +170 Noelani Kamelamela talk contribs (→R230) | ||||
|
|
09:52 | (Upload log) [Noelani Kamelamela (6×)] | |||
|
|
09:52 Noelani Kamelamela talk contribs uploaded File:R230pic.jpg | ||||
|
|
08:42 Noelani Kamelamela talk contribs uploaded File:R230pic.jpeg | ||||
|
|
08:32 Noelani Kamelamela talk contribs uploaded File:Syntax.jpg | ||||
|
|
08:31 Noelani Kamelamela talk contribs uploaded File:Droligoprocess.jpg | ||||
|
|
08:29 Noelani Kamelamela talk contribs uploaded File:Chamber.jpg | ||||
|
|
08:28 Noelani Kamelamela talk contribs uploaded File:DrOligo.jpg | ||||
2 May 2024
|
|
19:54 | Paper Microfluidic Device for Archiving Breast Epithelial Cells 5 changes history −389 [Xning098 (5×)] | |||
|
|
19:54 (cur | prev) −10 Xning098 talk contribs (→Diseases tested with neonatal heel pricks) | ||||
|
|
19:53 (cur | prev) −12 Xning098 talk contribs (→Results with Breast Epithelial Cells) | ||||
|
|
19:49 (cur | prev) 0 Xning098 talk contribs (→Current diagnostic tools) | ||||
|
|
19:45 (cur | prev) +45 Xning098 talk contribs (→Design Parameters) | ||||
|
|
19:40 (cur | prev) −412 Xning098 talk contribs (→Design Parameters) | ||||