Paper Microfluidic Device for Archiving Breast Epithelial Cells
History of Breast Cancer
460 B.C. - Hippocrates names breast cancer as karkinos, a Greek word for “crab”, because he described the tumors having crab leg shaped roots. [1]
200 A.D.- Galen considered breast cancer to be more dangerous and believed that it affected the whole body, which is why surgery was not an option then but rather medicinal therapies.[1]
The 1680s - Francois de la Boe Sylvius, a French physician, challenged the humoral theory of cancer, and debated that it was due to a chemical process transforming lymphatic fluids to have different properties. [1]
The 1730s - Claude-Deshais Gendron, another French physician, rejected Galen’s theory and challenged it stating breast cancer forms when nerve and glandular tissues interacted with lymph vessels. [1]
1757 - French physician, Henri Le Dran, suggested that surgery of the tumor and removing the infected lymph nodes may treat cancer. [1]
1894 - William Halsted developed radical mastectomy, where the surgeon removes the complete breast including the surrounding lymph nodes and chest muscles. [1]
1895 - George Beatson, a Scottish surgeon, discovered that the removal of one of his patient’s ovaries correlated to the reduction in tumor size in her breast. [1]
1940 - 1960 - Researchers hypothesized and established that ovarian and hormone suppression contributed to improving outcomes in breast cancer patients. [2]
1971- Total mastectomy, where the surgeon removed just breast tissue, showed more promising results compared to radical mastectomy in cancer patients. [3]
1974 - Chemotherapy drug, Adriamycin, showed tumor-shrinking abilities for severe breast cancer cases. [3]
1975 - Incorporation of a methyl group in a CpG motif is shown to influence gene expression, such as X-inactivation.[4]
1976 - Bernard Fisher suggested that breast cancer can metastasize. He published results of simpler breast-conserving surgical strategies followed by radiation and chemotherapy. [2]
1977 - Chemotherapy drug, Nolvadex, approved for treating severe cases of breast cancer.[5]
The late 1970s - Mammographies become more frequent; more cases earlier diagnosed at a more treatable stage. [6]
1983 - CpG regions of normal cells were found to be methylated, while cancer cell regions were not. [7]
1986 - Hypermethylation found to cause gene silencing for calcitonin, a marker for lung cancer. [7]
1989 - Hypermethylation shown to silencing tumor suppressor genes. [7]
The mid-1990s - Discovery of BRCA1 and BRCA2 genetic mutations contribute to increased cancer risks in patients. Bisphosphonates are also introduced to help reduce the complications associated with treatments. Sentinel lymph node biopsy to limit the amount of breast tissue removed during the mastectomy. [18]
2000 - Genetic sequencing allows for deeper insight in how to recognize various types of breast cancer. [1]
2004 - Genetic tests reveal significant predictors of cancer recurrence in patients. Such a test is OncoType DX. [1]
2006 - National Cancer Institute announces decreased mortality rates of breast cancer patients as a result of screening and treatments. [9]
2012 - 2013 - Trastuzumab emtansine (T-DM1; Kadcyla) shows promise in treating resistant HER2 positive cancers in clinical trials and became FDA approved in 2013. [1]
Breast Cancer: Overview
Breast cancer is the most frequent malignant disease among women, 1 in 8 will be diagnosed within her lifetime. In 2016 alone, there were 230,000 new cases and 40,000 deaths reported thus far. The two most common types of breast cancers are lobular carcinoma (LC) and ductal carcinoma (DC). LCs comprise of abnormal cells localized in the lobules of the breast where milk is produced; this type of cancer makes up 10% of those affected by this disease. DCs account for 80% of those who have breast cancer. These cancer cells originate in the breast ducts that transfer milk from the lobules to the nipple. [10]
Breast Cancer Genetics
Mutations in the genes BRCA1 and BRCA2 have been shown in a substantial proportion of high risk families for breast cancer. These genes are tumor suppressors that produce a protein to facilitate the repair of damaged DNA. Deletions in these genes cause the gene to no longer function, leading to damaged DNA and in some cases, breast cancer. Genetic inheritance of these damaged genes only accounts for around 10% of diagnosed breast cancer cases. [11]
Breast Cancer Epigenetics
Epigenetics are the heritable and reversible changes in gene expression not accompanied by an alteration in the DNA sequence. These modifications usually occur at the DNA level (DNA methylation) and chromatin level (chromatin remodeling) [Epigenetics of mammalian breast epithelial cells]. The important factors that are involved in the epigenetic regulation of all cancers are DNA methylation and histone modification involved in silencing tumor-suppressive genes [Figure 1]. An enzyme-driven chemical change, DNA methylation usually occurs at CpG islands. Normally, the majority of these islands are unmethylated; in a cancer cell, global levels of DNA methylation become reduced concurrently with increased DNA methylation of CpG islands. [11]
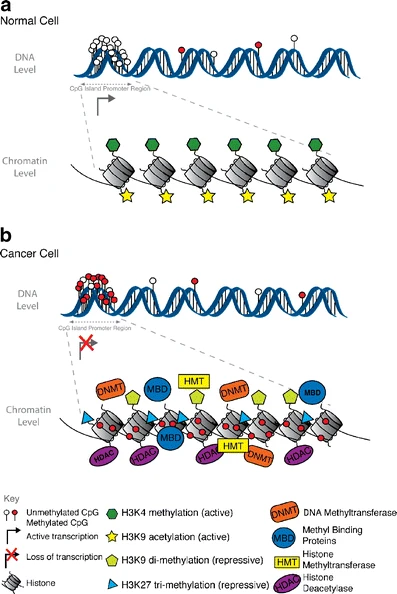
In breast cancer, it is the activation of oncogenes (a gene that predisposes normal cells to turn into cancerous cells) within mammalian breast epithelial cells (MBEC). There are over 100 different genes that are susceptible to methylation which makes identifying specific breast cancer genes a difficult feat. [12]
Current diagnostic tools
Early detection of breast cancer is essential to the survivor-ship of a patient that has been diagnosed with breast cancer. The following are just a few measures that individuals take in determining whether or not they have the disease.
Self-Examinations
Adult women are recommended to give themselves self-examinations at least once a month. The method consists of feeling around the breast and armpit area using the pads of her fingers from outside to center, taking note of any lumps or knots. The individual conducts this exam either in the shower, in front of a mirror, or lying down. Although not every lump in breast tissue indicates cancer, it allows for the patient to become familiar with their own breasts and be able to tell their healthcare professional of any abnormalities they found during the exercise. [13]
Mammograms
Mammograms are x-ray pictures of the breast that can be used to check for breast cancer in women that usually show no symptoms of the disease. There are two types of these x-rays: screening and diagnostic mammograms. Screening mammograms incorporate two of these images, one for each breast, and detect tumors that are unable to be located through touch (i.e. microcalcifications). Diagnostic mammograms are conducted after identifying a lump or other symptoms of the disease, and takes longer than a screening mammogram as more x-rays are involved with this screening. [14]
Duct Lavage
A duct lavage is a minimally invasive procedure usually performed in an outpatient facility or a doctors office, for women who present several risk factors. For the duct lavage, the doctor applies suction to the nipple, causing the ducts to open and release milk. A catheter is then inserted to the duct and saline solution is used to wash the duct and lobe. Epithelial cells are shed from the wash and drawn back up with the saline solution to be tested for cancer cells. This may be repeated for several ducts. While this procedure is minimally invasive, a doctor's visit is required and the procedure may be uncomfortable.
Biopsy
A biopsy is a small operation to collect tissue from the area in question, and is usually conducted if a mass appears on a mammogram. There are various biopsy techniques that a surgeon may use in order to collect a sample. The following are just a few of the least to most invasive biopsy procedures that are practiced [15]:
- Fine needle aspiration biopsy: hollow needle is inserted multiple directly into lump to collect tissue sample
- Core needle aspiration biopsy: hollow needle is inserted in breast about 3 to 6 times to collect tissue samples from suspicious area
- Vacuum-assisted breast biopsy: a probe is inserted into breast as a vacuum sucks up the breast tissue and a rotating cutting device removes the wanted sample
- Incisional biopsy: a scalpel is used to cut into the breast and a piece is removed for examination
- Excisional biopsy: a scalpel is used to remove the entire area of suspicious breast tissue
Current Complications associated with Breast Cancer Detection
Complications associated with breast cancer detection include invasiveness, late prognosis, false positives, and cost.
There is still ambiguity surrounding when a woman should get a mammogram; for low-income women that do not have insurance, there are only some clinics that offer free and low cost mammogram screenings. Not all lumps within the breast tissue are cancerous, which is why false positives may often lead to biopsies. Some types of breast cancers do not show obvious signs of development, making the location of the carcinomas difficult to pinpoint and the disease hard to diagnose.
In recent studies, delayed childbirth has had a strong correlation to the rise in breast cancer patients under 40. It is hypothesized that this is occurring because pregnancy may stimulate the growth of cells that may have already transformed from normal to cancerous cells as a result of high estrogen and progesterone levels. The hormonal and immunologic modifications in pregnancy support an environment for the proliferation of breast cancer cells. [16]
However, it is more difficult to diagnose these patients as current methods may pose as a threat to the woman and child, which is why most of pregnancy-associated breast cancer diagnoses occur postpartum. [17]
Possibilities in Breast Cancer Detection
Current Research Archiving Epithelial Cells from Breast Milk
Future state of breast cancer detection methods would be non-invasive and cheap equipment that are able to store tissue samples from patients. Research involving breast epithelial cells collected from breast milk shows promise of an easier way to detect the disease than collecting tissue samples through biopsies. [18]
Specifically relating to breastfeeding mothers, instead of going into a doctors office for an invasive procedure to check a woman for breast cancer, women can send in a small amount of breast milk which would be tested for epigenetic markers relating to a woman's individual risk for breast cancer. By using breast milk, the epithelial cells collected from the milk are from each lobe of the breast creating a more complete test of the entire breast. Research is currently underway relating DNA methylation on oncogenes genes to the development of breast cancer which would make this early detection test possible.
Results with Breast Epithelial Cells
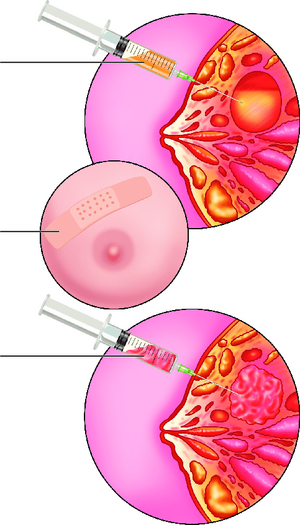
Research involving collecting breast milk to filter out breast epithelial cells has produced positive results allowing for the possibility to move away from tissue biopsy samples. The need for cheaper methods of breast cancer detection methods are more crucial than ever as the percentage of individuals developing this disease is increasing rapidly. At the Brazilian Biosciences National Laboratory in Campinas, Brazil, researchers utilize nipple aspirate fluid (NAF) from lactating females to conduct protein identification on Guthrie Cards using mass spectrometry. More than 30 major proteins were successfully identified using this method. [19] However, collecting NAF is still an invasive measure, requiring a needle to be inserted under the woman's nipple into the ductal lobes in order to collect the fluid. [Figure 2]
At the University of Massachusetts Amherst, Kathleen F. Arcaro, professor of Environmental Toxicology, and her team do similar work with regards to breast cancer but in a non-invasive manner. Collecting breast milk samples from two study groups, individuals with either biopsied or non-biopsied breasts who are able to lactate, their method includes storing the DNA onto Whatman FTA cards and extracting the DNA from the device. Unlike the previous example that uses NAF, the Arcaro lab collects breast milk from postpartum women. This experiment produced three results:
- The biopsied women had a slight increased chance of CpG promoter methylation in three specific genes that they were observing. [20]
- Two of the three genes observed showed different methylation patterns between the two sample groups. [20]
- A small number of the women in this study had significantly higher CpG promoter methylation. [20]
With these types of results, non-invasive and low cost methods seem more promising with the practice of paper microfluidics as potential DNA storage devices as part of the breast detection method. Also, these paper devices are much easier to transfer to research labs compared to milk samples held in bottles. This is important because more of these devices can be distributed and collected at a time, allowing for researchers to perform quicker analysis on the storage devices rather than worrying about their milk samples being destroyed in the delivery process.
Research Complications
Currently, the Arcaro Lab at UMASS Amherst collects breast milk from nursing mothers and later, if the mother develops breast cancer, tests the DNA obtained from the epithelial cells present in the milk for hypermethylation. Yet, the method of breast milk collection is inconvenient, costly, and requires a significant amount of milk from the mothers (~100 mL). Mothers must send in a bottle of milk (often without defining which breast the milk came from) which then must be transported back to the Arcaro lab in a bulky box refrigerating the milk. This process is a hassle not only for the mothers but for the researchers as well, thus motivation for a DNA storage device is high. Only a milliliter of milk is needed for the amount of DNA required to perform pyrosequencing, but larger amounts must be transported due to the low yield of DNA. More information about Professor Arcaro and her research can be found on her website
Paper Microfluidics
Paper microfluidics are low cost devices that use capillary flow to drive fluid through the device’s channels. Various paper microfluidic devices include pregnancy tests, immunoassays, and glucometer strips. These devices can be fabricated in many ways including wax printing patterns, through photolithography to create masks, and by laser cutting shapes into the paper itself. Since these devices are relatively cheap to manufacture, the low cost contributes to its ability to be mass produced.
Paper microfluidics being inexpensive, easy to assemble, and able to be mass produced is why these devices have been used as DNA storage agents in experimental methods to better improve breast cancer detection. The most common paper microfluidic devices for DNA storage are the Guthrie and Whatman FTA cards.
DNA Storage Cards
Guthrie Cards
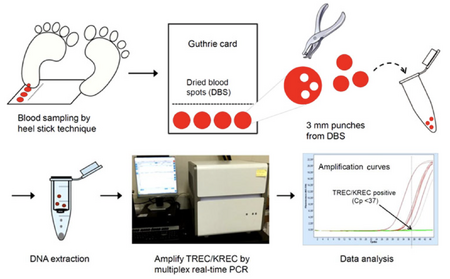
Invented by Robert Guthrie in the 1960s, these paper storage cards hold blood samples taken from newborns. Originally, Dr Robert Guthrie used dried blood spot (DBS) specimens for phenylalanine analysis to identify newborns with phenylketonuria. [22] In a neonatal heel prick, the newborn heel is pricked with a needle and the blood is directly applied to the Guthrie Card. When the fluid dries, small samples are taken from the card and placed in a buffer solution with bashing beads in order to extract the data [Figure 5]. [20] The diseases that the Guthrie Cards currently test for are listed in the following section.
Whatman FTA Cards
The Whatman FTA Card is another DNA storage device with similar functions to Guthrie Cards. Unlike the Guthrie Cards, these cards have coatings that allow for better stability of DNA in the DBS and longer storage life. [Figure 3] [23]

Diseases tested with neonatal heel pricks
The neonatal heel prick tests for various diseases depending on the country in which the child is born. Below is a list of some of the diseases the neonatal heel prick tests for: [21]
- Phenylketonuria
- Biotinidase Deficiency
- Congenital Adrenal Hyperplasia
- Congenital Hypothyroidism
- Congenital Toxoplasmosis
- Cystic Fibrosis
- Galactosemia
- Homocystinuria
- Maple Syrup Urine Disease
- Medium-Chain Acyl-CoA Dehydrogenase Deficiency
- Toxoplasma gondii IgM antibodies
PCR/Methylation Detection Strategy
Polymerase chain reaction (PCR) is a process used on either one or multiple copies of DNA. The mechanism behind PCR is based upon the ability of DNA polymerase to provide the complementary strand to a DNA template strand. The DNA template is the strand containing the target sequence. DNA polymerase is an enzyme that synthesizes the complementary strand to the target DNA sequence. It does this by using primers, short single stranded DNA strands that are complementary to the target sequence. DNA polymerase uses these primers to make DNA nucleotides for the new strand. The DNA template strand is the sequence of DNA that is amplified. PCR begins with the denaturation step, samples are heated to 94-96 degrees celsius in order to separate the target DNA into separate strands. The temperature is then reduced to 50-65 degrees celsius to allow the primers to anneal. The temperature is then raised to 72 degrees celsius for DNA polymerase to synthesize the new DNA strand at each primer’s attachment site.
DNA methylation is an epigenetic marker that controls whether gene expression is turned on or off. The mechanism behind DNA methylation occurs through the covalent transfer of C-5 position of the cytosine ring of DNA. The enzymes that perform the transfer are DNA methyltransferases.
The most common procedure to observe DNA methylation of a gene is called a bisulfite conversion. DNA is treated with bisulfite to remove an amine from the cytosine, converting it to uracil. However, cytosines that have been methylated are resistant to this process and are not converted to uracil. After this process PCR can then be performed, uracils will be amplified as thymines and cytosines will be amplified as cytosines showing a clear distinction between methylated DNA and non-methylated DNA.
In their studies, the Arcaro group found that samples that yielded less than 1 microgram of DNA were much more likely to result in an incomplete bisulfite conversion reaction and thus a failed pyrosequencing analysis. Samples containing at least 1 microgram of DNA were shown to result in better PCR results and a greater pyrosequencing success rate. [20]
Comparison: Neonatal heel prick diseases and Breast Cancer
The diseases that the neonatal heel prick tests for are usually developmental, genetic, and metabolic disorders in newborns. If the tests come back positive for the newborn having any of these maladies, the babies are able to receive immediate treatment.
For diseases such as breast cancer, it is harder to test for as it is a condition that takes longer to develop. Also, blood and buccal samples contain more cells than breast milk, which is why disorders are easier to detect using blood and buccal specimens. [24] However, in the case for postpartum women and understanding how breast cancer may be related to pregnancy, the use for these devices may still be applicable.
Potential of Paper Microfluidic Device for DNA Storage
Design Parameters
Important design parameters to consider in the design of any paper microfluidic device are the physical properties of the filter paper to be used, the viscous properties of the sample liquid, desired sample volume and the dimensions of the wax barrier. Two of the defining properties of filter papers are approximate pore diameter and paper thickness. Since filter papers are composed of randomly dispersed cellulose fibers, pore diameter measurements are approximated based upon particle retention tests rather than physical pore measurements. Choice of pore diameter is important in attempting to filter cells or particles using size exclusion or regulation of flow rate through the device.
Flow rate in filter paper may be approximated using Darcy’s law: [math]\displaystyle{ \mathrm{L} = \left(\frac{\gamma Dt}{4\eta}\right)^{1/2} }[/math] for fluid flow in porous media. [25] According to this model, flow rate is proportional to paper permeability, pore width, pore height and a pressure differential driving force and inversely proportional to fluid viscosity. If the sample fluid and volume are kept constant, pressure differential and viscosity will also remain constant. Thus, increasing pore diameter will increase flow rate as well by increasing both cross sectional area and permeability.
Another important consideration is the size of the hydrophobic wax barrier that is applied to the device. In the case that you wish to design a multilayered device, the surface area allowed for vertical flow through the device is an important limiting factor. Increasing this cross sectional area will increase absorption into the device. Before printing a wax design, the flow of melting wax into the device during barrier impregnation must be considered. The spreading of the wax is described by Washburn’s equation: [math]\displaystyle{ \mathrm{Q} = -\frac{kwh}{\mu}\frac{dp}{dx} }[/math]. [26] The length that the wax spreads from the printed design is proportional to the surface tension of the wax, the pore diameter of the paper and the amount of time the wax is allowed to flow. It is inversely proportional to the viscosity of the wax. Both surface tension and viscosity are properties of the wax that are dependent upon temperature, thus the penetration of the wax into the paper may be controlled by allowing the wax to melt at a set temperature and amount of time.
Complications
The overall goal of our paper based device is to be able to concentrate, store, and send through the mail epithelial cell samples from the breast milk of lactating mothers. Since the card is likely to be used in the presence of infants, it is highly important that the design contains no toxic materials.
One major point of concern in the study performed by the Arcaro group was the loss of DNA samples due to a failed pyrosequencing reaction. Average DNA yields for the reference sample group and the biopsy sample group were 2.6 and 0.671 micrograms respectively; thus, in the case that the pyrosequencing were to fail, not enough DNA was left over to perform a second bisulfite conversion and the sample is lost. As a result, 55.2% of samples for RASSF1, 40.3% for SFRP1 and 31.3% for GSTP1 were lost. [20] Therefore, it is our intention to design a device such that it will concentrate an epithelial cell sample and consistently provide a yield of at least 1 microgram of DNA.
Another point of interest with the previous study is the relatively small group of women from which breast milk samples were collected. Fresh milk samples were provided by 134 women from the area surrounding Amherst and, as stated previously, many of those samples were lost to failed pyrosequencing attempts. [20] Thus another goal of our paper device is the ability to send it through the mail, allowing for a much greater target population. Difficulties that arise when sending milk samples in the mail include wear and tear, degradation of DNA over time and microbial growth. The Whatman cards used previously provide a reliable model to follow in achieving this goal. Whatman cards are coated with both an antimicrobial detergent and uric acid (to prevent free radical degradation of DNA). Both of these coatings have been found to be effective in preserving the sample for several months.
Absorbent Breast Pad for DNA Storage
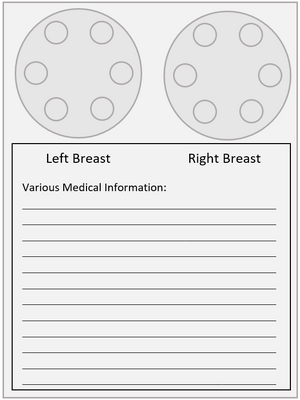
Device Design
Similar to the paper device, an absorbent breast pad used by nursing mothers is modified into a tissue transportation device in which mothers place a teaspoon of milk onto the pad from each breast. In order to concentrate the DNA in the sample to maximize DNA extraction, a barrier capable of confining the breast milk to a nickel sized area must be used. DNA is then extracted from the pads and tested for risk factors associated with breast cancer.
By using a breast pad, a product already designed to absorb breast milk, only slight modifications are needed in order to create a device capable of storing DNA. The pads' multiple layers facilitate absorption. Additionally since the device is an absorbent pad, unlike the paper device, the milk is absorbed immediately within seconds compared to hours for the paper device. By using the breast pad, which is commercially available and widely used, and slightly modifying them for a transportation device allows for the device to be low in cost and fairly easy to make. A special thanks to Kimberly-Clark for answering all of our questions and also providing materials for testing.
Similar to Guthrie Cards, the sample would need to be stored for a long period of time and still be able to be tested for valid information. Thus additional materials would need to be added to stabilize the DNA within the device. Thoughts on adding a positively charged polymer to the pad in order to bind the DNA and prevent the DNA from becoming damaged or broken down.
In order to test cells from each breast, the preliminary device design would include two pads clearly labeled and adhered to a paper describing the mothers information [Figure 6]. Known risk factors could be defined such as first degree family members, age, and race.
Device Complications
Of the few materials used to create a barrier on the pad, none were able to create a complete barrier to concentrate the milk in a small circle. The breast pads multiple layers present a challenge, since the barrier must span all three layers. Concentrating the milk to a smaller area of the pad would facilitate DNA extraction by maximizing the amount of DNA within the sample. Wax barriers proved to be unusable due to the fact that the wax cooled before it could penetrate all of the layers of the nursing pad. Wax printing could circumvent this as it uses higher temperatures, which could allow the wax to move through all layers. Silk screen printing could also be used to attempt to penetrate all layers of the nursing pad.
References
[1] Lukong, K. E. Understanding Breast Cancer – the Long and Winding Road. BBA Clinical 2017, 7, 64–77. DOI:https://doi.org/10.1016/j.bbacli.2017.01.001.
[2] Clarke, M. J. Ovarian Ablation in Breast Cancer, 1896 to 1998: Milestones along Hierarchy of Evidence from Case Report to Cochrane Review. BMJ 1998, 317 (7167), 1246–1248. DOI:https://doi.org/10.1136%2Fbmj.317.7167.1246.
[3] Fisher, B. Ten-Year Results of a Randomized Clinical Trial Comparing Radical Mastectomy and Total Mastectomy with or without Radiation. Plastic and Reconstructive Surgery 1985, 315 (11), 674-681. https://www.nejm.org/doi/abs/10.1056/NEJM198503143121102.
[4] Harrison, A.; Parle-Mcdermott, A. DNA Methylation: A Timeline of Methods and Applications. Frontiers in Genetics, 2011, 2. DOI:https://doi.org/10.3389/fgene.2011.00074
[5] Quirke, V. M. Tamoxifen from Failed Contraceptive Pill to Best-Selling Breast Cancer Medicine: A Case-Study in Pharmaceutical Innovation. Frontiers in Pharmacology 2017, 8. DOI:https://doi.org/10.3389/fphar.2017.00620.
[6] Nicosia, L.; Gnocchi, G.; Gorini, I.; Venturini, M.; Fontana, F.; Pesapane, F.; Abiuso, I.; Bozzini, A. C.; Pizzamiglio, M.; Latronico, A.; Abbate, F.; Meneghetti, L.; Battaglia, O.; Pellegrino, G.; Cassano, E. History of Mammography: Analysis of Breast Imaging Diagnostic Achievements over the Last Century. Healthcare 2023, 11 (11), 1596. DOI:https://doi.org/10.3390/healthcare11111596.
[7] Feinberg, A. P.; Tycko, B. The history of cancer epigenetics. Nature Reviews Cancer, 2004, 4(2), 143-153. DOI:https://doi.org/10.1038/nrc1279.
[8] Antoniou A, Pharoah PD, Narod S, Risch HA, Eyfjord JE, et al. Average risks of breast and ovarian cancer associated with BRCA1 or BRCA2 mutations detected in case series unselected for family history: a combined analysis of 22 studies. Am J Hum Genet. 2003, 72, 1117–1130. DOI: https://doi.org/10.1086/375033.
[9] National Cancer Institute (NCI). https://www.nih.gov/about-nih/what-we-do/nih-almanac/national-cancer-institute-nci (accessed 2024-03-10).
[10] American Cancer Society. BREAST CANCER. https://www.cancer.org/cancer/types/breast-cancer.html?utm_source=google&utm_medium=cpc&utm_campaign=Google+Grants+-+Cancer+Type+-+BMM&utm_term=breast%20cancer&gad_source=1&gclid=CjwKCAjw_e2wBhAEEiwAyFFFo-wfLMMB3-utEHU2LYyzwN80IerBRed39zSrYfeU3qB6bhh_YmnFfhoCVTgQAvD_BwE
[11] Mcpherson, K. ABC of breast diseases: Breast cancer---epidemiology, risk factors, and genetics. Bmj, 2000, 321(7261), 624-628. DOI:https://doi.org/10.1136/bmj.321.7261.624
[12] Hinshelwood, R. A.; Clark, S. J. Breast cancer epigenetics: normal human mammary epithelial cells as a model system.J Mol Med (Berl), 2008, 86(12), 1315-28. DOI:https://doi.org/10.1007/s00109-008-0386-3.
[13] Breast Self-Exam : The National Breast Cancer Foundation. [www.nationalbreastcancer.org, http://www.nationalbreastcancer.org/breast-self-exam]
[14] Mammograms. National Cancer Institute http://www.cancer.gov/types/breast/mammograms-fact-sheet#q8
[15] Biopsy. Breastcancer.org http://www.breastcancer.org/symptoms/testing/types/biopsy
[16] Callihan EB; Gao; Jindal S; Lyons TR. Postpartum diagnosis demonstrates a high risk for metastasis and merits an expanded definition of pregnancy-associated breast cancer. National Center for Biotechnology Information. Breast Cancer Res Treat, 2013, 138(2). DOI: https://doi.org/10.1007/s10549-013-2437-x.
[17] Borges, V. Management of the Patient With Postpartum Breast Cancer. Management of the Patient With Postpartum Breast Cancer http://www.cancernetwork.com/oncology-journal/management-patient-postpartum-breast-cancer
[18] Grote, A.; Abbas, M.; Linder, N.; Kreipe, H. H.; Lundin, J.; Feuerhake, F. Exploring the spatial dimension of estrogen and progesterone signaling: detection of nuclear labeling in lobular epithelial cells in normal mammary glands adjacent to breast cancer. Diagnostic Pathology. Diagn Pathol, 2014, 9. DOI:https://doi.org/10.1186/1746-1596-9-S1-S11.
[19] Delmonico, L.; Areias, V. R.; PINTO, R.O.D.R.I.G.O. É. S. A. R.; Matos, C. D. S.; Rosa, M. F. F.; Azevedo, C. M. D.; Alves, G. Protein identification from dried nipple aspirate fluid on Guthrie cards using mass spectrometry. Mol Med Rep, 2015, 12 (1). DOI:https://doi.org/10.3892/mmr.2015.3432
[20] Browne, E. P.; Punska, E. C.; Lenington, S.; Otis, C. N.; Anderton, D. L.; Arcaro, K.F. Increased promoter methylation in exfoliated breast epithelial cells in women with a previous breast biopsy. Epigenetics, 2011, 6(12), 1425-1435. DOI: https://doi.org/10.4161/epi.6.12.18280
[21] Hill, M.A. Embryology. Guthrie test. https://embryology.med.unsw.edu.au/embryology/index.php/guthrie_test. (accessed 2024-03-10).
[22] Cruickshank, M. N.; Pitt, J.; Craig, J. M. Going back to the future with Guthrie-powered epigenome-wide association studies. Genome Med, 2012, 4(10): 83. DOI: https://doi.org/10.1186/gm384
[23] The GENiSYSS Science of DNA Storage. [GENiSYSS, LLC, https://www.genisyss.com/science/]
[24] Choi, E.-H.; Lee, S. K.; Ihm, C.; Sohn, Y.-H. Rapid DNA Extraction from Dried Blood Spots on Filter Paper: Potential Applications in Biobanking. Osong Public Health and Res Perspect, 2014, 5(6): 351-7. DOI:https://doi.org/10.1016/j.phrp.2014.09.005
[25] Jang, I.; Song, S.; Facile and precise flow control for a paper-based microfluidic device through varying paper permeability. Lab on a Chip, 2015, 15, 3405-3412. DOI: https://doi.org/10.1039/C5LC00465A
[26] Carrilho, E.; Martinez, A. W.; Whitesides, G. M.; Understanding Wax Printing: A Simple Micropatterning Process for Paper-Based Microfluidics. Anal. Chem. 2009, 81, 7091-7095. DOI: https://doi.org/10.1021/ac901071p.
[27] Kathleen F. Arcaro. https://www.umass.edu/veterinary-animal-sciences/research-faculty/kathleen-f-arcaro (accessed 2024-03-10).
[28] Jain, Sudhir & Kaushik, Rohit. (2018). Diseases of Breast. https://www.researchgate.net/publication/327681516_Diseases_of_Breast
[29] Reliable DNA Extraction from Whatman® FTA® Cards. https://www.sigmaaldrich.com/US/en/technical-documents/protocol/genomics/dna-and-rna-purification/whatman-reliable-extraction-of-dna (accessed 2024-05-06).
[30]Son S.; Kang J.; Kim, J.M.; Sung,S.; Kim, Y.; Lee, H.; Kim, B.R.; Lee, Y.K.; Ko,S.Y.; Shin, S.M.; Kim Y. Pediatr Infect Vaccine. 2017 Dec;24(3):134-140. https://doi.org/10.14776/piv.2017.24.3.134
