Mia Huddleston Week 9
Electronic Notebook
Purpose
The purpose of this lab is to choose a topic for my HIV structure research project and begin research on the project.
Methods and Results
- When developing a question to research, Avery and I decided to look into how sequence changes of amino acids may be different for each progressor group.
- We decided, for simplicity, to only look at nonprogressor and rapid progressors to have a clear difference if possible.
- We also decided to only look at the first to last visit of each of the subjects we choose from both of the progressor groups.
- We then chose two subjects in each the nonprogressors and rapid progressors
- Subjects 3 and 10 and
- Subjects 12 and 13
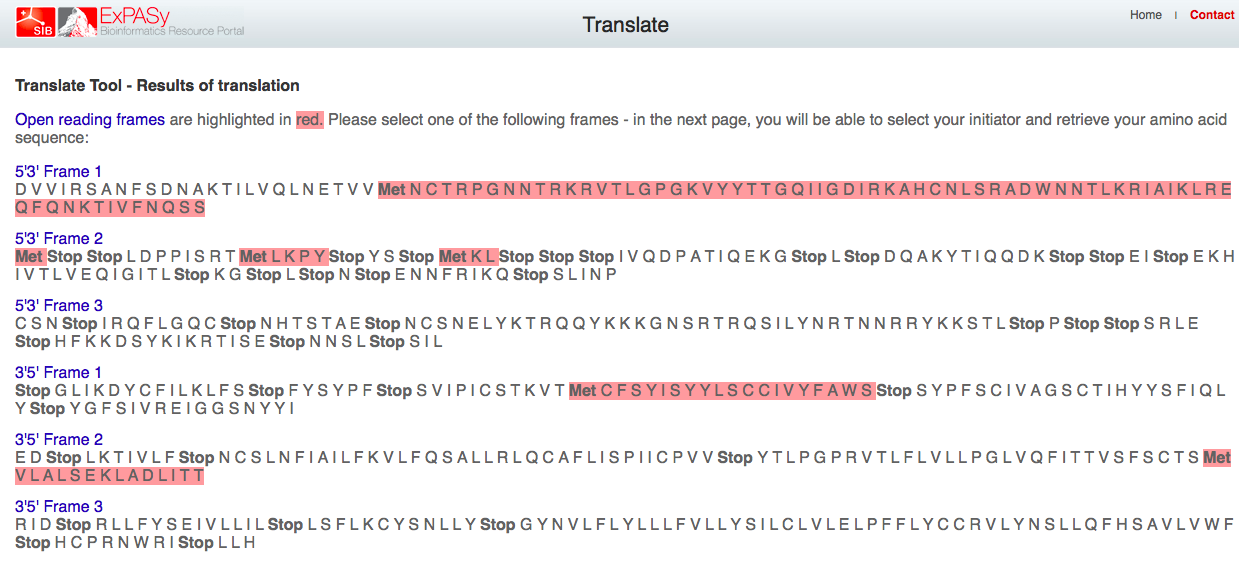
- Then, we converted our DNA sequences into protein sequences using the ExPASY Translate tool
- How do you know which of the six frames is the correct reading frame (without looking up the answer)?
- We know which of the reading frames is correct by which ones do not have stop condons in the middle of the reading frame since this sequence was taken from the middle of a complete open reading frame as seen in the picture below
- How do you know which of the six frames is the correct reading frame (without looking up the answer)?
- We then can find the rest of our sequence data from from the BEDROCK HIV Problem Space.
- To learn more about HIV gp120 envelope, we explored the UniProt Knowledgebase (UniProt KB)
- If you search on the keywords "HIV" and "gp120", in the main UniProt search field, how many results do you get?
- 206,278 results
- What types of information are provided about this protein in this database entry?
- This database gives information on the function, molecular function, and the biological process of this protein.
- If you search on the keywords "HIV" and "gp120", in the main UniProt search field, how many results do you get?
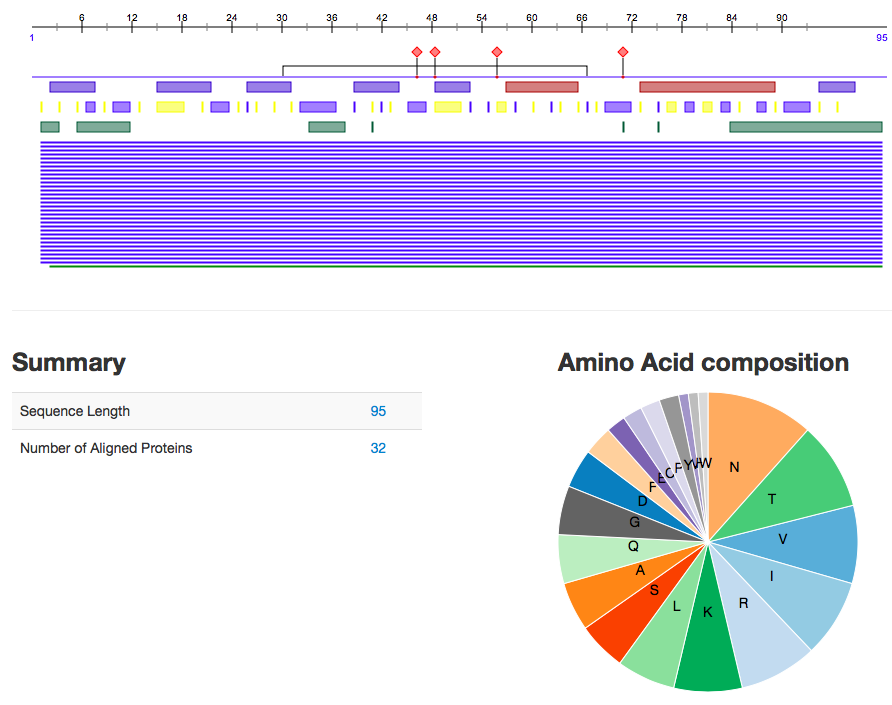
- We then used the PredictProtein server and imputed one of our amino acid sequences (S3V1-1) into the input field to predict the protein. A picture of the results can be seen below.
- How does this information relate to what is stored in the UniProt database?
- This gives you a lot of specific information about this sequence and the possible function just as UniProt did.
- How does this information relate to what is stored in the UniProt database?
HIV Structure Project:
- What is your question?
- Does diversity and divergence increase within the amino acid sequences over time more within the rapid progressors compared to non-progressors?
- Make a prediction about the answer to your question before you begin your analysis.
- Since divergence and diversity increases over time within the DNA sequences for all progressor types, and more so within the rapid progress ors, we would expect to see more diversity within the amino acid sequences over time within the rapid progressors.
- Which subjects, visits, and clones will you use to answer your question?
- For rapid progressors we are looking at subject 3 and 10's first and last visits and for non-progressors we are looking at subject 12 and 13's first and last visits. These were chosen based on similar number of visits.
Conclusion
In this assignment we found a research topic that relates changes in amino acids to changes in the progression of HIV-1. We decided to discuss how sequence changes of amino acids may be different for each progressor group. We chose four different subjects and will look at their first vs. last visit of non-progressors and rapid-progressors. We are still unsure of how to compare these visits, but will determine this next week.
Data and Files
Acknowledgements
Avery Vernon-Moore and I worked together to create the "Defining Your HIV Structure Research Project" portion. Dr. Dahlquist provided help in the writing and designing of our research project. While I worked with the people noted above, this individual journal entry was completed by me and not copied from another source. Mia Huddleston 19:51, 25 October 2016 (EDT)
References
- ExPASY Translate tool
- BEDROCK HIV Problem Space
- UniProt Knowledgebase (UniProt KB)
- Markham, R.B., Wang, W.C., Weisstein, A.E., Wang, Z., Munoz, A., Templeton, A., Margolick, J., Vlahov, D., Quinn, T., Farzadegan, H., & Yu, X.F. (1998). Patterns of HIV-1 evolution in individuals with differing rates of CD4 T cell decline. Proc Natl Acad Sci U S A. 95, 12568-12573. doi: 10.1073/pnas.95.21.12568
Useful links
User Page: Mia Huddleston
Bioinfomatics Lab: Fall 2016
Class Page: Bioinfomatics Laboratory, Fall 2016