Mia Huddleston Week 2
Electronic Lab Notebook
To begin this experiment we used the application Aipotu and followed the procedure under the biochemistry section to produce the following results. The purpose was to find a true-breeding purple flower from the known amino acid sequences of two green true-breeding flowers, a white true-breeding flower, and a red true-breeding flower.
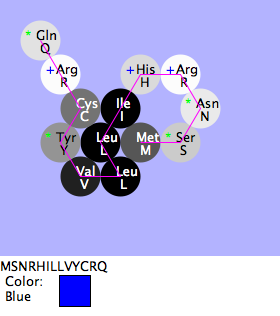
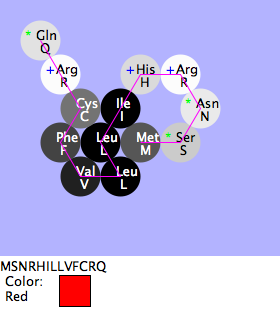
Mia, Shivum, Will: Blue vs. Red Flowers
- Purpose
- To find if the difference between blue and red is Phe vs. Tyr as the 10th amino acid sequence
- Methods and Result
- We compared the natural red flower with the blue that makes up the green-2. When compared every amino acid is the same except for one. The difference was only one amino acid: the 10th. When the 10th amino acid is Phe the color is red, when it is Tyr it is blue.
- Conclusion
- The hypothesis is correct, in this case the only difference between red and blue is one amino acid.
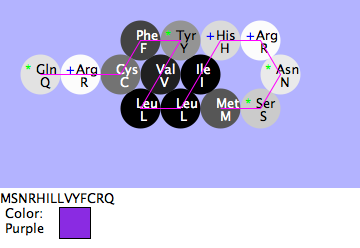
Mia, Shivum, Will: True-Breeding Purple Flower
- Purpose
- To find if when Phe is added to the blue flower's amino acid sequence next to Tyr, the blue flower will now become purple.
- Methods and Results
- I added Phe to the blue flower's amino acid sequence next to Tyr and folded this new amino acid sequence. I thought that the since the two were so similar except for one amino acid, but combined they created a purple flower, then when the amino acid sequence contained both amino acids naturally, then it would be purple. This resulted in a larger protein with a different structure that did end up being purple. Phe and Tyr have very similar structures with each having a benzene ring. When another benzene ring is added, the color can be changed.
- Conclusion
- The hypothesis was accurate, this addition created a true-breeding purple flower.
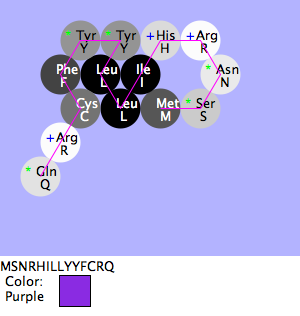
Mia, Shivum, Will: Second Purple Flower
- Purpose
- To know if you change the Val amino acid to a Tyr amino acid, then the flower will remain purple.
- Methods and Results
- Since Val and Tyr are very similar in their structure with both having benzene rings, I thought that they could possibly be interchangeable with their phenotype of purple even if their genotype has been changed. So, I placed the purple amino acid sequence in the upper panel and changed the Val to a Tyr in the sequence and folded this new sequence. This ended up confirming my hypothesis that the color remained purple, but it still created a new structure as seen in the image above.
- Conclusion
- The hypothesis was accurate, this change still resulted in a true-breeding purple flower.
- After finding these two flowers I experimented with other known amino acid sequences to find true-breeding yellow, orange, black, red, blue, and green flowers.
Final Conclusion
- After conducting these experiments I found that comparing two colored flowers that together combine to create a third colored flower, you could easily perceive a slight difference in amino acid sequences. Then, it was possible to create that third colored flower naturally by adding together these two amino acid sequences with both/all of the amino acids that separate the two. You could also change the amino acid sequence by switching out amino acids with similar structures and still result in the same colored flower as seen in experiment three.
Data and Files
- Images of all results can be seen in the electric lab notebook section.
Acknowledgements
I worked with my partners Shivum Desai and William Fuchs meeting in class working on the same prompt but on different computers. We had help from our professor, Dr. Dahlquist in class as well to complete this assignment. While I worked with the people noted above, this individual journal entry was completed by me and not copied from another source.
Mia Huddleston 19:15, 6 September 2016 (EDT)
References
The images and information used were gained from Aipotu using their application.
- Aipotu. (n.d.). Retrieved from http://aipotu.umb.edu
Resources used to create this page:
- Week 2 Assignment Page created by Dr. Kam D. Dahlquist
Useful links
User Page: Mia Huddleston
Bioinfomatics Lab: Fall 2016
Class Page: Bioinfomatics Laboratory, Fall 2016