Mia Huddleston Week 10
Electronic Notebook
Purpose
The purpose of this assignment is to continue to work on our projects and find the results to our hypothesis; we would expect to see more diversity within the amino acid sequences over time within the rapid progressors.
Methods and Results
- Download the structure file for the paper we read in journal club from the NCBI Structure Database.
- These files can be opened with the Cn3D software site that is installed on the computers in the lab (this software is free, so you can download it and use it at home, too.) Alternately, you may choose to use the Star Biochem program to do this portion of your work. Answer the following:
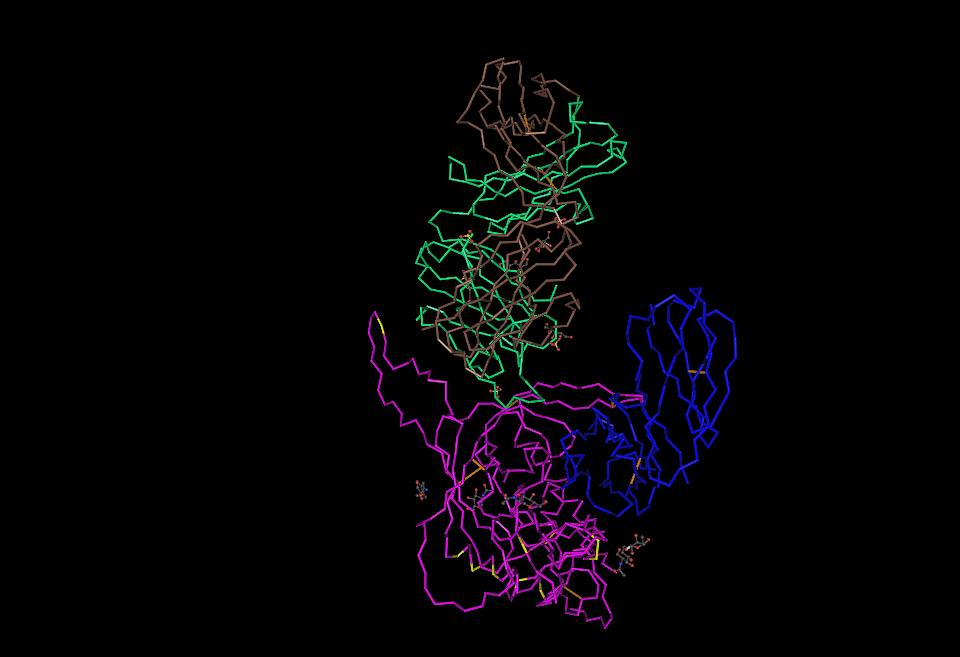
- Find the N-terminus and C-terminus of each polypeptide tertiary structure.
- Locate all the secondary structure elements. Do these match the predictions made by the PredictProtein server?
- Yes, these do match the predictions
- Locate the V3 region and figure out the location of the Markham et al. (1998) sequences in the structure.
- I found the V3 region using the sequences that we have from the markham data starting at "evv"
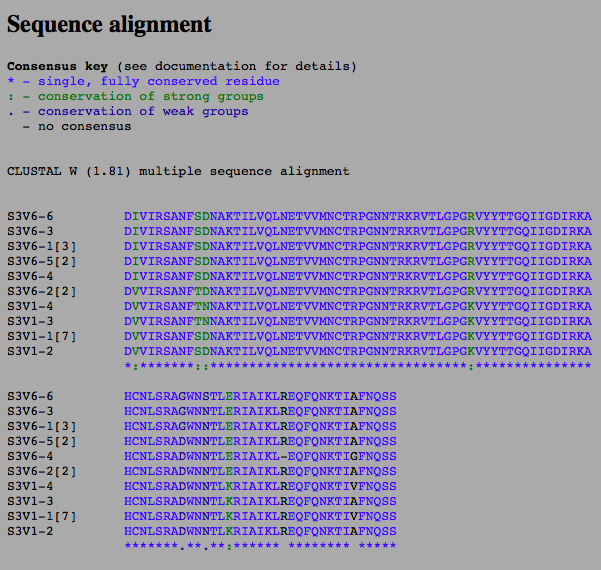
- Perform a multiple sequence alignment on the protein sequences that you and your partner are analyzing for your project using the Biology Workbench.
- Are there more or fewer differences between the sequences when you look at the DNA sequences versus the protein sequences?
- There are more differences between the DNA sequence than the protein sequence.
- How do you account for this?
- This is because a change in the sequence may not end up changing the amino acid
- Are there more or fewer differences between the sequences when you look at the DNA sequences versus the protein sequences?
- Once you have oriented yourself, analyze whether the amino acid changes that you see in the multiple sequence alignment would affect the 3D structure and explain why you think this.
- We found, comparing what the change in aa, if the change would be large or small depending on the mutation and from there if the change would effect the structure
- I found sequence alignments between the first and last visits of subjects 3 and 10 using the Biology Workbench and rooted trees for each. Subject 3's alignment can be seen bellow. Avery found 12 and 13's alignments.
- We then compared our sequences and the changes seen in the alignment of the V3 region to the sequence found by Kwong et. al in their paper.
- Bellow, subject 10's placement of aa that have been altered are all shown in yellow of the pink region.
- This was mapped for all subjects. In subject 10, it can be seen that there is one mutation on the V3 loop.
- We then also compared which mutations would result in big changes in function and which may not.
- Then, we also performed clustldist using the biology workbench to determine the amount of diversity between each visit
- Subject 12: 0.010
- Subject 13: 0.021
- Subject 10: 0.137
- Subject 3: 0.010 (Only four clones in last visit)
HIV Structure Project:
- What is your question?
- Does diversity and divergence increase within the amino acid sequences over time more within the rapid progressors compared to non-progressors?
- Make a prediction about the answer to your question before you begin your analysis.
- Since divergence and diversity increases over time within the DNA sequences for all progressor types, and more so within the rapid progress ors, we would expect to see more diversity within the amino acid sequences over time within the rapid progressors.
- Which subjects, visits, and clones will you use to answer your question?
- For rapid progressors we are looking at subject 3 and 10's first and last visits and for non-progressors we are looking at subject 12 and 13's first and last visits. These were chosen based on similar number of visits.
Conclusion
In conclusion, we found sequence alignments, rooted trees, and clustldists for each of the four subjects looking at each of their first and last visits using the biology workbench. There appeared to be more diversity within the rapid progressors, especially for subject 10, but p-values were not found. There was not as much of a difference for subject 3, but we concluded that this could be because there are less total clones to analyze for subject 3. In the future we would want to look at more subjects with more clones and more visits. We also looked at where the mutations occurred in gp120 and what kind of mutations occurred using the Kwon et al. figure.
Data and Files
Presentation
Acknowledgements
Avery Vernon-Moore and I worked together to create the "Defining Your HIV Structure Research Project" portion. We also met at our house on Monday and Wednesday to finish our powerpoint which we completed together using google docs and Avery uploaded. Dr. Dahlquist provided help in the writing and designing of our research project. While I worked with the people noted above, this individual journal entry was completed by me and not copied from another source. Mia Huddleston 18:33, 7 November 2016 (EST)
References
- ExPASY Translate tool
- BEDROCK HIV Problem Space
- UniProt Knowledgebase (UniProt KB)
- Markham, R.B., Wang, W.C., Weisstein, A.E., Wang, Z., Munoz, A., Templeton, A., Margolick, J., Vlahov, D., Quinn, T., Farzadegan, H., & Yu, X.F. (1998). Patterns of HIV-1 evolution in individuals with differing rates of CD4 T cell decline. Proc Natl Acad Sci U S A. 95, 12568-12573. doi: 10.1073/pnas.95.21.12568
Useful links
User Page: Mia Huddleston
Bioinfomatics Lab: Fall 2016
Class Page: Bioinfomatics Laboratory, Fall 2016