Kvescio Week 14 Assignment
From OpenWetWare
Jump to navigationJump to search
Weekly Assignments
- user:kvescio ~ Instructions: BIOL368/S20:Week 1
- kvescio Week 2 Assignment ~ Instructions: BIOL368/S20:Week 2
- kvescio Week 3 Assignment ~ Instructions: BIOL368/S20:Week 3
- kvescio Week 4 Assignment ~ Instructions: BIOL368/S20:Week 4
- kvescio Week 5 Assignment ~ Instructions: BIOL368/S20:Week 5
- kvescio Week 6 Assignment ~ Instructions: BIOL368/S20:Week 6
- kvescio Week 8 Assignment ~ Instructions: BIOL368/S20:Week 8
- kvescio Week 10 Assignment ~ Instructions: BIOL368/S20:Week 10
- kvescio Week 11 Assignment ~ Instructions: BIOL368/S20:Week 11
- kvescio Week 13 Assignment ~ Instructions: BIOL368/S20:Week 13
- kvescio Week 14 Assignment ~ Instructions: BIOL368/S20:Week 14
Class Journals
- BIOL368/S20:Class Journal Week 1
- BIOL368/S20:Class Journal Week 2
- BIOL368/S20:Class Journal Week 3
- BIOL368/S20:Class Journal Week 4
- BIOL368/S20:Class Journal Week 5
- BIOL368/S20:Class Journal Week 6
- BIOL368/S20:Class Journal Week 10
- BIOL368/S20:Class Journal Week 11
- BIOL368/S20:Class Journal Week 13
- BIOL368/S20:Class Journal Week 14
Purpose
- The Purpose of the Final Project was to evaluate the SARS-CoV-02 virus and how it binds to ACE2 receptors in species other than humans, then to predict whether or not those species will be able to be infected with COVID-19
Combined Methods/Results
Choosing topic and evaluating species with ACE2
- Discussed topic with group and decided to research based on Wan et al. paper.
- Looked at species that also have the ACE2 receptor.
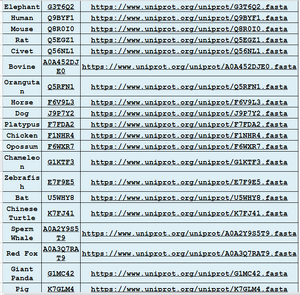
- Chose 19 different species from UniProt Knowledgebase (UniProt KB)
- Considered which species would be different from each other and interesting to research.
- Chosen species were:
- Platypus
- Opossum
- Elephant
- Sperm Whale
- Horse
- Dog
- Red Fox
- Chicken
- Turtle
- Chameleon
- Zebrafish
- Obtained FASTA sequences from all 19 species.
- Table of sequences created by group, shown below:
Creating phylogenetic trees
- Input FASTA sequences in Phylogeny.fr.
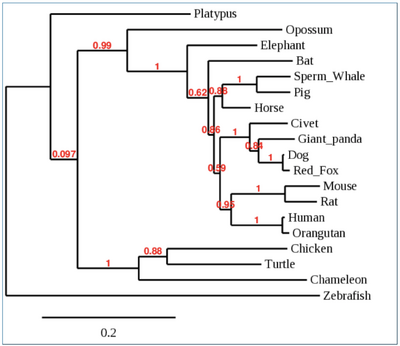
- First phylogenetic tree was created to evaluate all 19 species and their entire sequences.
- Phylogenetic tree shown below:
- No patterns were observed in first phylogenetic tree, other than the fact that the Platypus was a outlier.
- Human and orangutan grouped together, as well as mouse and rat.
- Group discussed importance of last 20 residues in sequence.
- Last 20 residues contain the binding region of the ACE2 receptor.
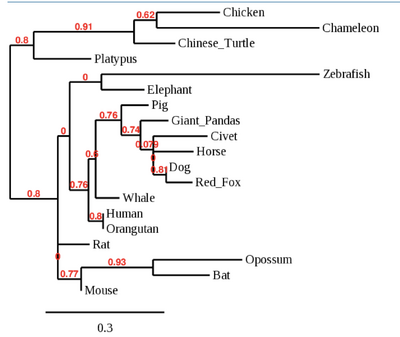
- Made a phylogenetic tree of 19 species, looking at just the binding region.
- Tree shown below:
Multiple Sequence Alignment
- Group creating multiple sequence alignment of 19 species.
- Used CLUSTAL format.
- Specifically looked at last 20 residues in sequence alignment.
3D Structure of SARS-CoV-02 and ACE2
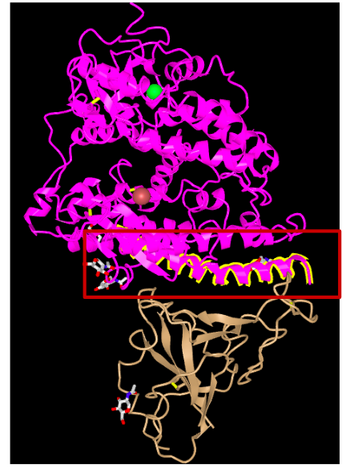
- Looked at iCn3D view of binding interaction between virus and receptor.
- Went to Protein Data Bank and searched "2AJF" obtained from Wan et al.(2020) paper.
- Analyzed 3D structure and discovered which part was the binding between the virus and ACE2 receptor.
- Image shown below highlights the part of the structure that is the interaction:
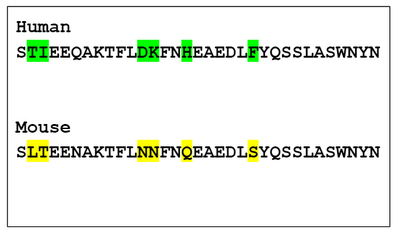
Residue differences in Mouse ACE2 ⍶ helix
- Chose to compare human ACE2 binding helix to the mouse ACE2 helix, since it is known that a mouse's receptor cannot bind to the novel virus.
- Mouse and human sequences are compared in image below:
- The results showed that the mouse and human had 6 differences in the binding part of the ACE2 receptor.
- Image highlights the 6 different residues.
- For remainder of project, those 6 residue locations were deemed the most important in evaluating binding properties in other species.
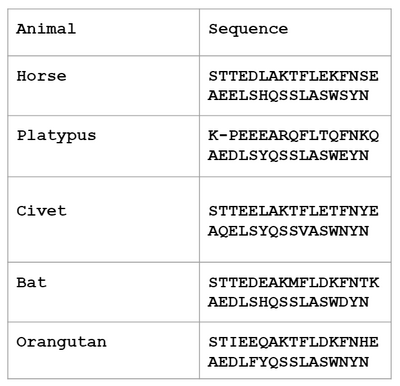
Comparing 5 species ACE2 receptor
- Group decided to narrow down project to 5 specific animals rather than 19.
- Animals chosen were:
- Horse
- Platypus
- Civet
- Bat
- Orangutan
- Those 5 animals were chosen due to their similarities and differences.
- The Horse and the Civet are similar, and since it is known that other cats can get the virus, we wanted to look closer at the civet.
- Civet was also chosen because of its ability to bind to ACE2 receptor in the SARS-CoV-01 virus.
- Platypus was chosen as an outlier, because there are no similarities whatsoever.
- Bat was chosen because it is the origin of the SARS viruses.
- Orangutan was chosen because it has the exact same ACE2 receptor sequence as the human.
- Table of 5 species and their sequences, shown below:
Looking at each animal more closely
Horse
- Some residues were preserved in the Horse.
- I→ T= nonpolar to polar
- D→ E= both - charged
- H→ S= + charged to polar
- F→ S= nonpolar to polar
Platypus
- Platypus was different from Human in all residues.
- T→ - = loss of polar A.A.
- I → P = both hydrophobic, change of shape
- D→T = - charged to uncharged
- K→ Q = + charged to uncharged
- H→ K = both + charged, change of shape
- F→ S = hydrophobic to polar
Civet
- Only one residue was preserved in Civet
- I→ T = nonpolar to polar
- D→ E = both - charged
- K→ T = + charge to polar
- H→ Y = + charge to polar
- F→ S = nonpolar to polar
Bat
- Half of the residues are conserved in Bat
- I→ T = nonpolar to polar
- H→ T = + charged to polar
- F→ S = nonpolar to polar
Orangutan
- Orangutan is the exact same as human.
Predictions about COVID-19
- Which species can bind to SARS-CoV-02, based on their ACE2 receptor?
- Group standards suggest that animals with 3 or more similar residues (based on the 6 we investigated) could bind to the virus.
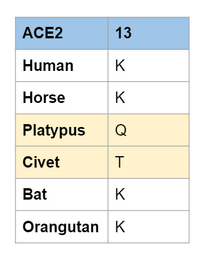
- Based on information from Wan et al. (2020), we concluded that residue location 13 was the most important to look at.
- Position 13 is a hotspot for binding interactions due to interactions that form a salt bridge.
- Image below shows location 13 in the species we evaluated.
- Based on this information, we predict that the Platypus and the Civet are not able to bind to the virus.
- Due to the fact that the Horse and Bat have many similarities in the ACE2 receptor to that of the human, and most importantly, at location 13, we predict that they are able to bind to the novel virus.
Discussion
- Outside research states that Horses can be infected with Equine Coronavirus.
- Fact supports our prediction that Horses' ACE2 receptor can bind to SARS-CoV-02, since it is also a Coronavirus.
- SARS-CoV-1 was able to bind to civets’ ACE2 receptor.
- Although our results suggest that civets cannot bind to the novel virus, it is a known fact that they were able to bind the previous SARS virus.
- However, civets were only able to bind to SARS-CoV-01 after it was mutated.
- SARS-CoV-02 has not mutated yet, so this aligns with our results predicting that civets' ACE2 cannot bind to the novel virus.
Future Research
- More research needs to be done on SARS-CoV-02 in general, and on the binding process in the ACE2 receptor.
- Future research after our findings would include:
- Studying how certain species can block SARS-CoV-2 using their ACE2 receptor.
- This would be beneficial in creating a treatment/vaccine for the current pandemic.
- Analyze why SARS-CoV-1 was able to mutate and bind to the ACE2 receptor in civets, but not SARS-CoV-2.
- Confirm whether or not civets can bind to the novel virus, and continue research if the virus mutates.
- Studying how certain species can block SARS-CoV-2 using their ACE2 receptor.
Data and Files
- https://openwetware.org/wiki/File:Uniprotfastamking.PNG
- https://openwetware.org/wiki/File:PhylotreeKV.png
- https://openwetware.org/wiki/File:Phylotree2KV.png
- https://openwetware.org/wiki/File:3dviewerace2mking.PNG
- https://openwetware.org/wiki/File:Humanmouseace2mking.PNG
- https://openwetware.org/wiki/File:5speciesmodifiedhorsemking.PNG
- https://openwetware.org/wiki/File:Horsemking.PNG
- https://openwetware.org/wiki/File:Platypusmking.PNG
- https://openwetware.org/wiki/File:Civetmking.PNG
- https://openwetware.org/wiki/File:Batmking.PNG
- https://openwetware.org/wiki/File:Orangutanmking.PNG
- https://openwetware.org/wiki/File:Position13mking.PNG
Scientific Conclusion
- After evaluating the 5 species, Horse, Platypus, Civet, Bat and Orangutan, it is observed that the Horse, Bat and Orangutan have the most similarities to the human ACE2 receptor. The criteria of having 3 or more similar residues, out of the 6 most important, were true for those species (Horse, Bat, Orangutan). They all also had the same as the human, at residue 13. Due to fitting those criteria, we predict that the Horse, Bat and Orangutan ACE2 receptor will be able to bind to the SARS-CoV-02 virus. However, the Platypus had no similarities to the human ACE2 receptor, so it is highly unlikely that it would be able to bid the the novel virus. Lastly, the civet had only one conserved residue, and most importantly, it was not conserved at residue 13. For those reasons, we predict that the civet ACE2 is not able to bind to SARS-CoV-02.
Final Powerpoint PDF
File:Maddy,karina,maya,lizzy.pdf
Acknowledgements
- I spoke with my partners Maddy, Lizzy, and Maya via text and zoom, and we worked on the assignment together.
- I asked Maddy if I could use some of the images she had already uploaded onto Openwetware.
- I used the images Maddy had uploaded for this assignment, since we all worked on it together and discussed it.
- I had a session with Lizzy on zoom to work on our sections of the powerpoint together.
- I copied the protocol from Week 14 for this assignment.
- Except for what is noted above, this individual journal entry was completed by me and not copied from another source.
(Kvescio (talk) 23:35, 29 April 2020 (PDT))
References
- OpenWetWare. (2020). BIOL368/S20:Week 14. Retrieved April 29, 2020, from https://openwetware.org/wiki/BIOL368/S20:Week_14
- Phylogeny.fr: Home. (n.d.). Retrieved April 29, 2020, from http://www.phylogeny.fr/
- Home - ORFfinder - NCBI. (n.d.). Retrieved April 29, 2020, from https://www.ncbi.nlm.nih.gov/orffinder/
- NCBI Protein Domains and Macromolecular Structures. (n.d.). Retrieved April 29, 2020, from https://www.ncbi.nlm.nih.gov/Structure/index.shtml
- RCSB Protein Data Bank. (n.d.). Homepage. Retrieved April 29, 2020, from https://www.rcsb.org/
- UniProtKB (2020). National Center for Biotechnology Information (NCBI). Retrieved April 29, 2020, from https://www.uniprot.org
- Wan, Y., Shang, J., Graham, R., Baric, R. S., & Li, F. (2020). Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. Journal of virology, 94(7). DOI: 10.1128/JVI.00127-20.