Kvescio Week 6 Assignment
Weekly Assignments
- user:kvescio ~ Instructions: BIOL368/S20:Week 1
- kvescio Week 2 Assignment ~ Instructions: BIOL368/S20:Week 2
- kvescio Week 3 Assignment ~ Instructions: BIOL368/S20:Week 3
- kvescio Week 4 Assignment ~ Instructions: BIOL368/S20:Week 4
- kvescio Week 5 Assignment ~ Instructions: BIOL368/S20:Week 5
- kvescio Week 6 Assignment ~ Instructions: BIOL368/S20:Week 6
- kvescio Week 8 Assignment ~ Instructions: BIOL368/S20:Week 8
- kvescio Week 10 Assignment ~ Instructions: BIOL368/S20:Week 10
- kvescio Week 11 Assignment ~ Instructions: BIOL368/S20:Week 11
- kvescio Week 13 Assignment ~ Instructions: BIOL368/S20:Week 13
- kvescio Week 14 Assignment ~ Instructions: BIOL368/S20:Week 14
Class Journals
- BIOL368/S20:Class Journal Week 1
- BIOL368/S20:Class Journal Week 2
- BIOL368/S20:Class Journal Week 3
- BIOL368/S20:Class Journal Week 4
- BIOL368/S20:Class Journal Week 5
- BIOL368/S20:Class Journal Week 6
- BIOL368/S20:Class Journal Week 10
- BIOL368/S20:Class Journal Week 11
- BIOL368/S20:Class Journal Week 13
- BIOL368/S20:Class Journal Week 14
Purpose
Purpose of this week's assignment was to go over research question, do our own research about the HIV virus in relation to Markham's study, and come up with our own conclusions based on data.
Research Question
Are the interim visits important to record and analyze the decline of CD4 T cells in order to accurately map the progression of HIV evolution?
Combined Methods/Results
Statistical Analysis of CD4 T cells count and sequenced data
- Looked at the data from Markham et al. 1998 data table
- Compared visits from each subject
- Saw that each subject had a first and last visit, however, not all visits had been sequenced.
- Chose the first visit from each subject and the Last Visit that was sequenced.
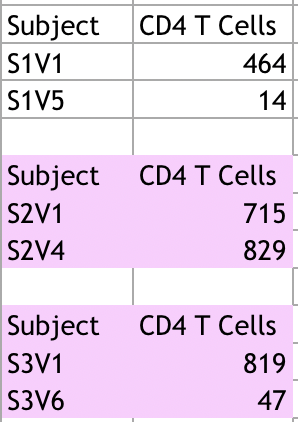
- Example of how we put information on Excel:
- Those subjects highlighted in pink were subjects who had future visits, however, had not been sequenced at those visits.
Graphed and calculated the slope in order to classify the subjects
- Took data from first and last sequenced visits.
- Put data on excel and created linear graphs.
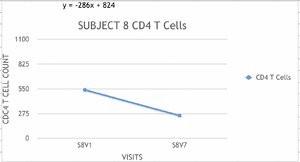
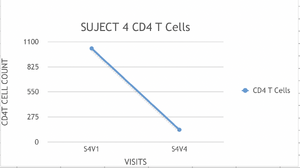
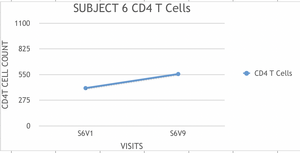
- Examples of graphs from subjects in different progressor categories:
- Got Equation of graph and slope of each graph.
- Compared the graphs of each subject, and their slopes.
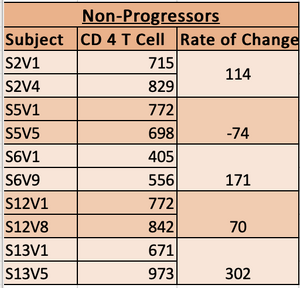
- Classified each subject into group of Non-Progressor, Moderate Progressor or Rapid Progressor.
- Decided classification rules.
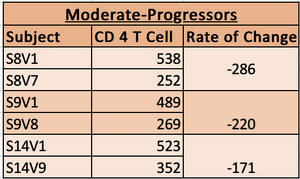
- Subjects who's slopes were anywhere from -100 to -300 were classified as moderate progressors.
- Subjects who's slopes were greater than -100 were classified as non-progressors.
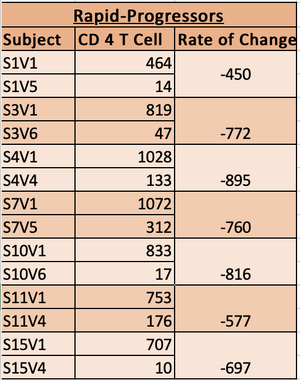
- Subjects who's slopes were less than -350 were classified as rapid Progressors.
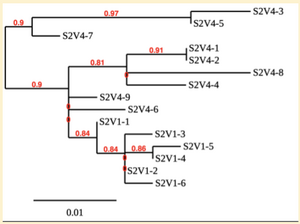
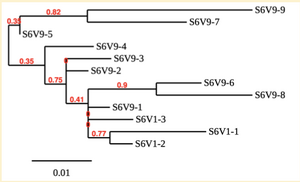
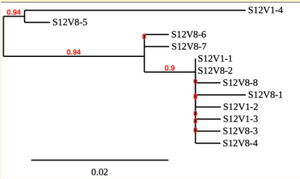
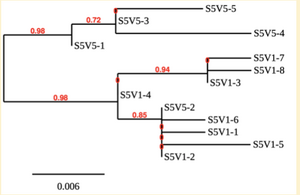
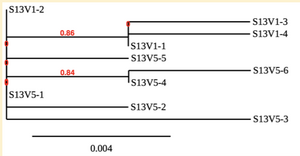
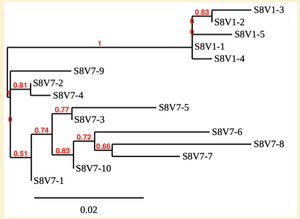
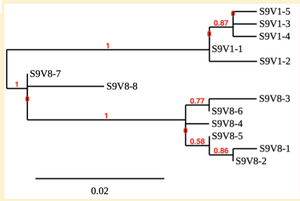
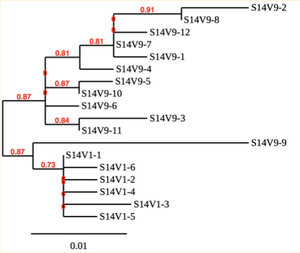
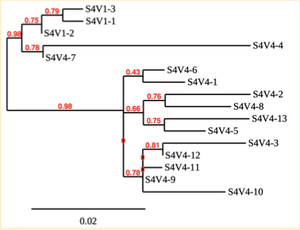
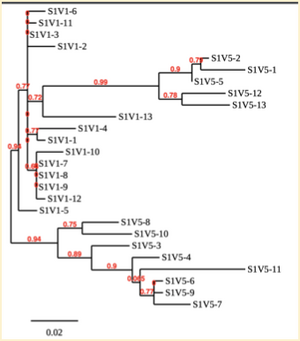
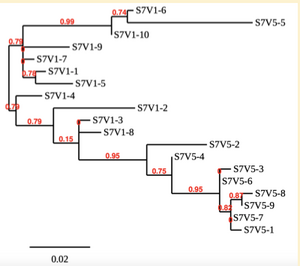
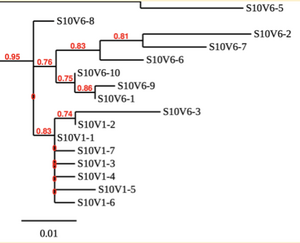
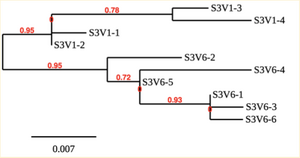
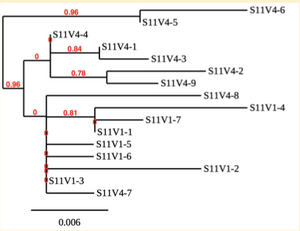
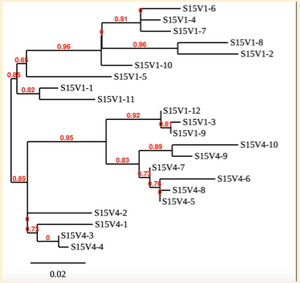
Created phylogenetic trees for each subject
- Created phylogenetic trees on Phylogeny.fr for each subject using clones of first and last visits.
Analysis of the change in CD4 T-Cell count from first visit to last visit in each of the progressor types.
- Classification of each subject based on their slope.
- Slope from -100 to -300 were moderate progressors
- Slope > -100 = non-progressors
- Slope < -350 = rapid progressors
Compared and contrasted our classification with Markham’s
- Our classification was the same as Markham's except for subjects 5, 6 and 7.
- Markham classified subjects 5 6 and 7 each as moderate progressors.
- We classified subjects 5 and 6 as non progressors and subject 7 as a rapid progressor.
Data and Files
- 13 tree
- subject 2 tree
- subject 6 tree
- subject 12 tree
- subject 5 tree
- subject 8 tree
- subject 9 tree
- subject 14 tree
- subect 4 tree
- subject 1 tree
- subject 7 tree
- subject 10 tree
- subject 3 tree
- subject 11 tree
- subject 15 tree
- non progressor chart
- moderate progressor chart
- rapid progressor chart
- File:Change in CD4 T-Cell count and HIV evolution from first to last visit in injection drug users.pdf
Scientific Conclusion
CD4 T cell decline is directly proportional to the evolution of HIV. In this project, it was discovered that Non-Progressors favored absence of change, while maintaining stable CD4 T cell counts. Such was shown in the phylogenetic trees of Non-Progressors because they showed less divergence and diversity. Progressors favored against absence of change, revealing a relationship between mutation and decline in CD4 T cell counts. The phylogenetic trees of Moderate and Rapid Progressors showed increase in diversity and divergence. The exclusion of analysis of interim visits did not affect the results of overall CD4 T cell decline. Therefore, lack of studying the interim visits was not detrimental to experiment. However, the lack of sequencing in last visits of certain subjects made it difficult to get full analysis. For future research, it would be important to sequence the subjects at every visit, so that there is full data for each of them. It would also be beneficial to record when the subjects stopped participating in the study.
Acknowledgments
- I copied directions and protocol from Week 6.
- I worked with Lizzy Urbina and Sahil in class and outside of class to work on project and create presentation.
- Copied and pasted nucleotide sequences for Part 1 from PubMed.
- Made phylogenetic trees and sequence Clustal forms via Phylogeny.fr
- Used data from Week 5 assignment page Nucleotide Sequence Data
- Communicated with Kam Dahlquist about research question.
- Except for what is noted above, this individual journal entry was completed by me and not copied from another source.
Kvescio (talk) 19:29, 26 February 2020 (PST)
References
- OpenWetWare. (2020). BIOL368/S20:Week 6. Retrieved February 26, 2020, from https://openwetware.org/wiki/BIOL368/S20:Week_6
- Nucleotide Sequences. (n.d.). Retrieved February 20, 2020, from http://bioquest.org/bedrock/problem_spaces/hiv/nucleotide_sequences.php
- http://www.phylogeny.fr/
- Markham, R.B., Wang, W.C., Weisstein, A.E., Wang, Z., Munoz, A., Templeton, A., Margolick, J., Vlahov, D., Quinn, T., Farzadegan, H., & Yu, X.F. (1998). Patterns of HIV-1 evolution in individuals with differing rates of CD4 T cell decline. Proc Natl Acad Sci U S A. 95, 12568-12573. doi: 10.1073/pnas.95.21.12568 (PubMed ID: 9770526)