BIOL368/S20:Week 5
This journal entry is due on Thursday, February 20, at 12:01am Pacific time.
Overview
The learning objectives for this assignment are:
- Learn several ways to quantify and visualize sequence similarity and difference.
- Ask your own questions and develop your own hypotheses to explain patterns in the Markham et al. (1998) dataset.
Individual Journal Assignment
Homework Partners
- You will be expected to consult with your partner, in order to complete the assignment.
- Each partner must submit his or her own work as the individual journal entry (direct copies of each other's work is not allowed).
- You must give the details of the interaction with your partner in the Acknowledgments section of your journal assignment.
- Homework partners for this week are:
- Christina and Annika
- Jenny and Nick
- Drew and Jack
- Karina, Lizzy and Sahil
- Madeleine and Maya
- Carolyn and Nathan
Format and Content Checklist
- Store this journal entry as "username Week 5" (i.e., this is the text to place between the square brackets when you link to this page).
- Write something in the summary field each time you save an edit. You are aiming for 100%.
- Invoke the template that you made as part of the Week 1 assignment on your individual page. Your template should contain:
- Purpose: a statement of the scientific purpose of the assignment. Note that this is different than the learning objective stated on the assignment page. What science will be discovered by completing this assignment?
- Combined Methods/Results (Electronic Lab Notebook): documentation of your workflow for this exercise. It should include:
- The protocol you followed in enough detail for someone else to be able to conduct the same investigation. There should be enough detail provided so that you or another person could re-do it based solely on your notebook. You may copy protocol instructions on your page and modify them as to what you actually did, as long as you provide appropriate attribution.
- Answers to any specific questions posed in the exercise.
- Screenshots and images to document your answers.
- Data and files: links to all data and files used and generated.
- Files left on the Desktop or My Documents or Downloads folders on the Seaver 120 computers will be deleted upon restart of the computers. Files stored on the
T:drive will be saved. However, it is not a good idea to trust that they will be there when you next use the computer. - Thus, it is a critical skill for data and computer literacy to back-up your data and files in at least two ways:
- Upload the files to this wiki.
- Upload the files to Box.
- Back them up on your personal flash drive.
- References to data and files should be made within the methods and results section. In addition to these inline links, create a "Data and Files" section of your notebook to make a list of the files generated in this exercise.
- Files left on the Desktop or My Documents or Downloads folders on the Seaver 120 computers will be deleted upon restart of the computers. Files stored on the
- Scientific Conclusion: a summary statement of the main result of exercise/research. It should mirror the purpose. Length should be 2-3 sentences, up to a paragraph.
- Acknowledgments section (see Week 1 assignment for more details.)
- You must acknowledge your homework partner with whom you worked, giving details of the nature of the collaboration. You should include when and how you met and what content you worked on together.
- Acknowledge anyone else you worked with who was not your assigned partner. This could be the instructor, the TA, other students in the class, or even other students or faculty outside of the class.
- If you copied
wiki syntaxor a particular style from another wiki page, acknowledge that here. Provide the user name of the original page, if possible, and provide a link to the page from which you copied the syntax or style. - If you copied any part of the assignment or protocol and then modified it, acknowledge that here and also include a formal citation in the Reference section.
- You must also include this statement:
- "Except for what is noted above, this individual journal entry was completed by me and not copied from another source."
- Sign your Acknowledgments section with your wiki signature (four tildes,
~~~~).
- References section (see Week 1 assignment for more details.)
- Use the APA format.
- Cite this assignment page.
- Cite any protocols that you copied and modified (this must also be noted in the Acknowledgments section).
- Cite any other methods, software, websites, data, facts, images, documents (including the scientific literature) that was used to generate content on your page.
- Do not include extraneous references that you do not cite or use on your page.
Exploring HIV Evolution: An Opportunity for Research
Acknowledgments
This exercise has been copied and modified with permission from this source:
- Donovan, S.S. & Weisstein, A.E. (2017). Exploring HIV Evolution. NIBLSE Incubator: Exploring HIV Evolution, /groups/ni_hivevolution, (Version 2.0). QUBES Educational Resources. Accessed February 13, 2020 from https://qubeshub.org/qubesresources/publications/24/2
Instructions for using Phylogeny.fr were modified from an unpublished lab exercise:
- Weisstein, A.E. (n.d.) Lab Module 3: Phylogenetics Part 2. Building phylogenetic trees using sequence data, personal communication.
Resources, Links, Data, and Files
- BEDROCK HIV Problem Space
- Dereeper, A., Guignon, V., Blanc, G., Audic, S., Buffet, S., Chevenet, F., ... & Claverie, J. M. (2008). Phylogeny. fr: robust phylogenetic analysis for the non-specialist. Nucleic acids research, 36(suppl_2), W465-W469. doi: 10.1093/nar/gkn180
- Vlahov, D., Anthony, J.C., Munoz, A., Margolick, J., Nelson, K.E., Celentano, D.D., Solomon, L., Polk, B.F. (1991). The ALIVE study, a longitudinal study of HIV-1 infection in intravenous drug users: description of methods and characteristics of participants. NIDA Res Monogr 109, 75-100. (PubMed), (Table of contents and link to download).
- HIV Database at Los Alamos National Laboratory
- Cells Alive HIV Tutorial
- Centers for Disease Control and Prevention- HIV Fact Sheet
- The Body: the HIV/AIDS Resource
- Avert: Global information and education on HIV and AIDS
Overview
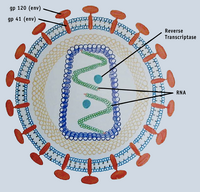
Human Immunodeficiency Virus (HIV), like other retroviruses, has a much higher mutation rate than is typically found in organisms that do not go through reverse transcription (the copying of RNA into DNA). Mansky and Temin (1995) estimated the rate of point mutations in HIV to be 3 x 10-5 errors per base per replication cycle. The impact of HIV as a pathogen is due in large part to this high mutation rate which, among other things, causes the surface proteins to change and avoid normal immune detection and suppression. A great deal has been learned about the evolution of HIV because it is relatively easy to sample the rapidly changing population of viruses within an infected individual and look at the patterns of molecular change over time.
In this activity you will study aspects of sequence evolution by working with a set of HIV sequence data from 15 different subjects (Markham, et al., 1998). You will first learn about the dataset, then study the possible sources of HIV for these subjects, and then design and pursue your own research project.
Basic HIV biology such as life history characteristics of the virus and its interactions with the human immune system are not discussed here but are important background for placing these exercises into a broader biological and clinical context. A few suggested resources for reviewing basic HIV biology are provided in the reference section.
An orientation to the HIV sequence data
Activity 1: Looking at the NCBI Resources and HIV sequence data
Part 1: PubMed
The NCBI provides access to a variety of databases including PubMed (a literature database) and GenBank (a nucleic acid sequence database). This federally funded research resource is very useful in part because the databases are linked together. This means that finding information in one area will allow you to look up related information in other areas. You should find the PubMed record for the Markham et al. article and then follow the links option for that record to find all the nucleotide sequences associated with that article.
NCBI website URL: http://www.ncbi.nlm.nih.gov
Original paper: Markham, R.B., Wang, W.C., Weisstein, A.E., Wang, Z., Munoz, A., Templeton, A., Margolick, J., Vlahov, D., Quinn, T., Farzadegan, H., & Yu, X.F. (1998). Patterns of HIV-1 evolution in individuals with differing rates of CD4 T cell decline. Proc Natl Acad Sci U S A. 95, 12568-12573. doi: 10.1073/pnas.95.21.12568 (PubMed ID: 9770526)
- How did you search for the PubMed entry?
- What other ways might you have searched?
- What other types of related information are available?
Part 2: GenBank
In this section you will take a closer look at a GenBank record and the type of data that is stored there. Once you reach the nucleic acid data associated with the Markham et al. paper you will see that there are a variety of different ways to view the data.
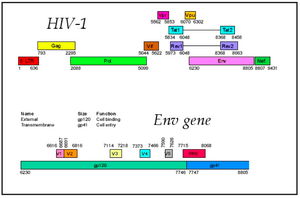
The data you will be working with is coded to help you recognize its source. While all of the data are HIV sequences, each sequence is identified based on the subject it was taken from, the visit during which it was collected, and its clone number. Thus, each sequence has a code like S4V2-4 that can be read as subject 4, visit 2, 4th clone. Each clone is a unique sequence collected during a particular visit. Over 600 different HIV sequences were identified in these 15 subjects and published electronically in GenBank. Choose one of the GenBank records and view both the full record and the FASTA formatted sequence.
- What was the accession number of the sequence you chose?
- Which subject of the study was that HIV sequence from?
- Which section of the record contains information about who the HIV was collected from?
- Download several (4 to 6) sequences in FASTA format to your local hard drive.
- Click the "Send to" link in the upper right of the page. Select "Complete Record", "File" as the Destination, and "FASTA" as the format. Click the "Create File" button. Be careful to remember where you put the file and what you name it so that you can find it later.
- Open the file that you saved with a word processor to confirm that you have the sequences and that they are in the FASTA format. In the FASTA format each sequence is preceded by a label which begins with the greater than sign (>). For example:
>AF016760.1 HIV-1 subject 1 visit 1 clone 1 from USA, envelope glycoprotein V3 region (env) gene, partial cds GAGGTAGTAATTAGATCCGAAAATTTCACGAACAATGCTAAAATCATAATAGTACAGCTGAATGAATCTG TAGAAATTAATTGTACAAGACCCAACAACAATACAAGAAAAAGTATACATATAGGACCAGGTAGAGCATT TTATACAACAGGAGACATAATAGGAGATATAAGACAAGCATATTGTAACATTAGTAGAGCAGAATGGGAT AACACTTTAAAACAGATAGTTATAAAATTAAGAGAACACTTTGGGAATAAAACAATAGTCTTTAATCACT CTTCA
Part 3: Introduction to Phylogeny.fr
In order to analyze sequence data we will use the Phylogeny.fr, a free, simple to use web service dedicated to reconstructing and analysing phylogenetic relationships between molecular sequences.
- In your browser, go to the website www.phylogeny.fr. Scroll down on the page to the section labeled ‘Phylogeny analysis’, and click on the text ‘One Click’.
- Click in the large text field labeled ‘Upload your set of sequences in FASTA, EMBL, or NEXUS format’. Use Ctrl-V to paste your sequences here, then click the “Submit” button.
- You will see a page named ‘Alignment results’. After your alignment is complete, you will see a new page named ‘Phylogeny results’. Finally, you will see a page named ‘Tree rendering results’. You will come back to these pages later. For now, find the numbered tabs located just beneath the text ‘One Click Mode’, and click on the tab labeled ‘3. Alignment’.
- Within the alignment, individual positions are color-coded to indicate their conservation, or how similar the sequences are to each other at that position. Blue highlighting indicates high conservation (i.e., the sequences are identical or at least very similar), while gray highlighting indicates lower conservation and white highlighting indicates little if any conservation.
- Near the bottom of the page, under ‘Outputs’, click on ‘Alignment in Clustal format’. This will display your alignment in a text-only format in which each position’s conservation is indicated by a symbol underneath the alignment block (“*” for invariant, “:” for highly conserved, “.” for weakly conserved, and a space for not conserved). Copy and paste this entire alignment into your individual wiki.
- Hint: After you paste it in, add a single space character to the beginning of each line. This will show the text in a fixed width font and preserve the alignment of letters.
- Now go back and click on the tab ‘6. Tree Rendering’, and you will see a phylogenetic tree, similar to Figures 3 & 4 of the Markham et al. (1998).
- On this tree, horizontal lines (branches) represent individual evolutionary lineages. By contrast, vertical lines (splits) represent mutation events, and the vertical length of each split is drawn purely for visual clarity with no biological meaning. The left-most split is called the root of the tree, and represents a hypothesis about the most recent common ancestor (MRCA) of the sequences within your tree.
- The length of each branch represents the percentage change in nucleotide sequence occurring along that branch, relative to the scale bar shown at the bottom of the tree. The scale bar will be a number between 0 and 1 and can be reinterpreted as a percent. for example. 0.05 would be 5%. The tree may also contain support values for each clade; shown in red on the branches, also expressed as a number between 0 and 1. 0.05 would be 5%. In general, a higher support value indicates a higher statistical confidence in a particular clade.
- Download the image in PNG format, giving the image a unique filename.
- Upload and display this image on your individual wiki page.
- Compare the tree to the multiple sequence alignment. See if you can relate the differences in the sequences to the topology of the tree diagram.
- Note that the sequence labels on your tree will be the accession numbers for those sequence records. With just the accession numbers to describe the sequences, it may be difficult to think about the biological basis for the comparisons you just performed. Going forward, instead of downloading sequences directly from GenBank, sequence files labeled as they were in the Markham et al. (1998) paper are provided.
Activity 2: Looking at the sources of HIV across subjects
For this activity you will work with HIV sequence data collected from 15 individuals from an injection drug using population in Baltimore. The goal of the study will be to determine if the HIV isolated from particular subgroups of subjects derives from a common source. In order to approach this question you will need to characterize the populations of HIV within an individual and quantify the differences between individuals. The work will be distributed among groups to make the research more manageable. You can then combine your own results with those from other groups to try to draw some conclusions about the entire population of subjects.
Most of your analyses will involve building multiple sequence alignments and distance-based unrooted trees. A multiple sequence alignment involves lining up several sequences so that comparisons can be made across sequences for each nucleotide position. Differences between related sequences can then be interpreted as nucleotide substitutions. Building alignments sometimes involves inserting gaps that represent areas where an insertion or deletion has taken place in one or more of the sequences. You will be using the Phylogeny.fr tool to build your alignments and distance-based unrooted trees.
Part 1: Looking at clustering across subjects
- Download the files
- Together these 2 files contain 97 sequences from the 15 subjects’ first visits (see Table 1). The sequence labels for these data include information about the subject, visit and clone number for each sequence.
This research was a prospective study, meaning that the subjects were receiving regular HIV screening and the first visit for this dataset indicates the first time they tested HIV positive. Remember, each clone consists of one or more copies of the virus representing a unique sequence in the V3 region of the env gene.
| Subject | Number of clones at visit 1 |
|---|---|
| 1 | 13 |
| 2 | 6 |
| 3 | 4 |
| 4 | 3 |
| 5 | 8 |
| 6 | 3 |
| 7 | 10 |
| 8 | 5 |
| 9 | 5 |
| 10 | 7 |
| 11 | 7 |
| 12 | 4 |
| 13 | 4 |
| 14 | 6 |
| 15 | 12 |
- As a preliminary analysis you should generate a multiple sequence alignment and distance tree for 12 of these sequences (3 clones from each of 4 subjects).
- Document your work by showing your sequence alignment and tree on your individual wiki page.
- Use the data table below (Table 2) to keep a record of the data you analyzed.
| Subject | Clone # |
- In tracing the branches between any two terminal nodes, or sequences, the total length traced reflects the genetic distance between those sequences. We can use the genetic distances as a first-order approximation of the evolutionary relationships between the sequences. The greater the genetic distance, the longer the time since they shared a common ancestor. However, because it is an unrooted tree we can’t make any inferences about the direction of the evolutionary change. An important feature of unrooted trees are long internal branches. Long internal branches (i.e., branches that don’t connect to terminal nodes) separate clusters of sequences. For example, Figure 5a suggests that sequences 1 and 2 are much more similar to each other than either of them is to sequences 3 or 4. On the other hand, Figure 5b does not provide strong evidence for grouping sequences.
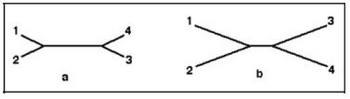
- Do the clones from each subject cluster together?
- Do some subjects' clones show more diversity than others?
- Do some of the subjects cluster together?
- Write a brief description of your tree and how you interpret the clustering pattern with respect to the similarities and potential evolutionary relationships between subjects' HIV sequences.
Part 2: Quantifying diversity within and between subjects
This section will introduce several additional ways to quantify sequence similarity and difference. These measures will help you design and interpret your research in the next section.
Table 1 illustrates that different subjects have different numbers of clones in their initial samples. This is one crude way to quantify the diversity of the HIV within that individual at that time, assuming that each subject’s sample contains approximately the same total number of viruses. The S statistic can also be used to quantify the diversity of sequence in a population. The value of S is simply the number of positions that vary, or are not identical, across all the sequences in an alignment.
- Select all the clones from one subject and align them.
- Data can be downloaded from: Nucleotide Sequence Data
- Document your work by showing your sequence alignment on your individual wiki page.
- From the alignment calculate S by counting the number of positions where there is at least one nucleotide difference across the collection of clones. Enter your data into Table 3 below (make a copy of this table and include it on your wiki page).
- Run the same analysis for a second and third subject and record your results in Table 3.
| Subject | Number of Clones | S | Theta |
|---|---|---|---|
- Next you will calculate θ (theta), which is an estimate of the average pairwise genetic distance. It is an estimate because it does not take into account the frequency of the different clones in the sample. θ is given by the formula:
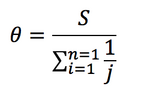
- where S is the statistic you just calculated above and the denominator of θ is a partial harmonic sum. For a sample of N = 5 the denominator would be calculated by 1 + 1/2 + 1/3 + 1/4 = 25/12. The harmonic sum is used because we do not expect the number of differences to increase linearly with the number of sequences analyzed.
- Calculate θ for the three subjects you chose to work with and enter the results in the data table.
Activity 3: Defining your HIV evolution research project
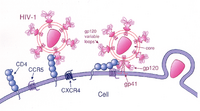
The data you used in Activity 2 was just a fraction of the data available from this study. The Markham et al. research followed this populations of 15 subjects over time and continued to sample and characterize their HIV populations at 6-month intervals for up to four years. In addition to studying the HIV population, the researchers estimated the immunological function of the subjects at each visit by measuring the number of CD4 T-cells in their blood samples. The CD4 T-cell count is an indicator of the health of the immune system because it represents a class of immune cells that are attacked by the HIV virus. A CD4 count below 200 cells/microliter is one of the diagnostic indicators used for defining Acquired Immune Deficiency Syndrome (AIDS).
There is a table summarizing the entire dataset on the BEDROCK website. There are also files containing all the data from all the visits made by each subject. These data files are saved in FASTA format with labels like those used in the dataset you used in Activity 2.
- Review the data summary table for the complete Markham et al. data set: Data table README
- Discuss with your partner/trio any patterns you see in the summary data table and possible research questions related to those patterns.
For this portion of your journal assignment, your electronic lab notebook entry should contain the answers to the questions below, which will define your HIV Evolution research project for the next couple of weeks. For this portion of the assignment only, both partners may have the same text on their individual journal pages (since you will develop the answers together for your joint project).
- What is your question?
- Make a prediction (hypothesis) about the answer to your question before you begin your analysis.
- Which subjects, visits, and clones will you use to answer your question?
- You should choose a combination of subjects, visits, and clones that will add up to approximately 50 sequences. You will need about that many sequences to answer a reasonably complex question. However, using much more than that will add too much complexity to the project.
- Data files are available at the BEDROCK site.
- Justify why you chose the subjects, visits, and clones you did.
Once you have your question, hypothesis, and data you will use, you should then move on to answering your question (see the Week 6 Assignment). Your electronic notebook should contain your notes, methods, results, and interpretations as you carry out your project. You should document as you work, taking your notes on the wiki as much as possible. Post data, figures, screenshots, to support your project. You can post files that are in progress; remember, you can upload a new version of the file and the wiki will automatically link to the new version (while keeping the old).
References
- Dereeper, A., Guignon, V., Blanc, G., Audic, S., Buffet, S., Chevenet, F., ... & Claverie, J. M. (2008). Phylogeny. fr: robust phylogenetic analysis for the non-specialist. Nucleic acids research, 36(suppl_2), W465-W469. doi: 10.1093/nar/gkn180
- Donovan, S.S. & Weisstein, A.E. (2017). Exploring HIV Evolution. NIBLSE Incubator: Exploring HIV Evolution, /groups/ni_hivevolution, (Version 2.0). QUBES Educational Resources. Accessed February 13, 2020 from https://qubeshub.org/qubesresources/publications/24/2
- Kwong, P. D., R. Wyatt, J. Robinson, R. W. Sweet, J. Sodroski, W. A. Hendrickson (1998). Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature, 393(6686):648-659. doi: 10.1038/31405 (PubMed ID: 9641677)
- Mansky, L. M., & Temin, H. M. (1995). Lower in vivo mutation rate of human immunodeficiency virus type 1 than that predicted from the fidelity of purified reverse transcriptase. Journal of virology, 69(8), 5087-5094. (PubMed ID: 7541846)
- Markham, R.B., Wang, W.C., Weisstein, A.E., Wang, Z., Munoz, A., Templeton, A., Margolick, J., Vlahov, D., Quinn, T., Farzadegan, H., & Yu, X.F. (1998). Patterns of HIV-1 evolution in individuals with differing rates of CD4 T cell decline. Proc Natl Acad Sci U S A. 95, 12568-12573. doi: 10.1073/pnas.95.21.12568 (PubMed ID: 98445411)
Shared Journal Assignment
- Compose your journal entry in the shared Class Journal Week 5 page. If this page does not exist yet, go ahead and create it (congratulations on getting in first :) )
- Create a header with your name, and then answer the questions in your own section of the page.
- You do not need to invoke your template on the class journal page.
- Any 'Acknowledgments and References you need to make should go in the appropriate sections on your individual journal page.
- Sign your portion of the journal with the standard wiki signature shortcut (
~~~~). - Add the category "BIOL368/S20" to the end of the wiki page (if someone has not already done so).
Reflect
- What is your comfort level when working with the bioinformatics tools during the in-class activity? What would increase your comfort level?
- Did the in-class discussion of the journal article enhance your understanding of the article? Why or why not?
- Have your views about what it means to do original research in biology changed as a result of discussing this article? Why or why not?
- What are the characteristics that you look for in a team member for a successful group project?