Kvescio Week 13 Assignment
From OpenWetWare
Jump to navigationJump to search
Weekly Assignments
- user:kvescio ~ Instructions: BIOL368/S20:Week 1
- kvescio Week 2 Assignment ~ Instructions: BIOL368/S20:Week 2
- kvescio Week 3 Assignment ~ Instructions: BIOL368/S20:Week 3
- kvescio Week 4 Assignment ~ Instructions: BIOL368/S20:Week 4
- kvescio Week 5 Assignment ~ Instructions: BIOL368/S20:Week 5
- kvescio Week 6 Assignment ~ Instructions: BIOL368/S20:Week 6
- kvescio Week 8 Assignment ~ Instructions: BIOL368/S20:Week 8
- kvescio Week 10 Assignment ~ Instructions: BIOL368/S20:Week 10
- kvescio Week 11 Assignment ~ Instructions: BIOL368/S20:Week 11
- kvescio Week 13 Assignment ~ Instructions: BIOL368/S20:Week 13
- kvescio Week 14 Assignment ~ Instructions: BIOL368/S20:Week 14
Class Journals
- BIOL368/S20:Class Journal Week 1
- BIOL368/S20:Class Journal Week 2
- BIOL368/S20:Class Journal Week 3
- BIOL368/S20:Class Journal Week 4
- BIOL368/S20:Class Journal Week 5
- BIOL368/S20:Class Journal Week 6
- BIOL368/S20:Class Journal Week 10
- BIOL368/S20:Class Journal Week 11
- BIOL368/S20:Class Journal Week 13
- BIOL368/S20:Class Journal Week 14
Purpose
- The purpose of Week 13 assignment is to be able to navigate different databases and tools in order to analyze the structure of SARS-CoV-02 and the ACE2 binding receptor. The different tools should allow one to investigate the inner structure of the virus and receptor.
Combined Methods/Results
Spike Protein Structure Exercise
Part 1
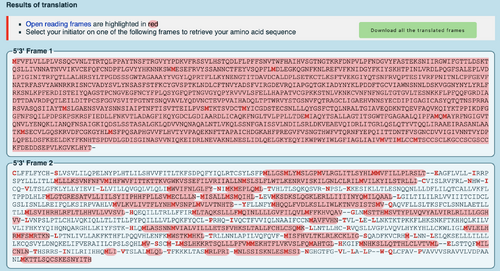
- DNA sequence of the SARS-CoV-2 S protein, found on Week 13 was converted to protein sequence using the ExPASY Translate tool
- photo of results:
- Frame 1 is the correct reading frame because it is the only one highlighted entirely in red, and it starts with "M"
- Checked my results with NCBI protein record, and concluded that they are correct.
Part 2
- By going to UniProt Knowledgebase (UniProt KB), I searched SARS-CoV
- I got 833 results from the beach.
- The result with the accession number "P59594" had information about the spike protein for SARS-CoV
- The database included information about the Spike proteins S1, S2, and S2'.
- It also included information about molecular function and biological process.
Part 3
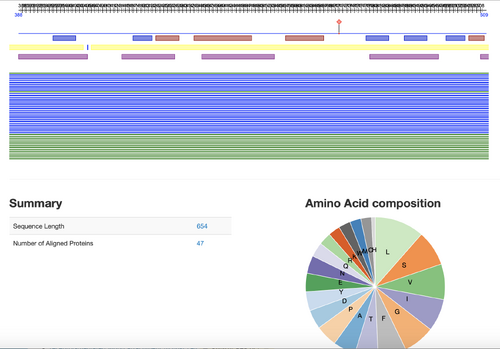
- Used PredictProtein server to analyze the SARS-CoV-2 spike protein.
- Sequence from article by Yan et. al. (2020).
- Downloaded sequence FASTA file from 6M17. Downloaded image below:
- Put sequence into PredictProtein server
- Took over 24 hours to get sequence back.
- Results from website:
- On Predict protein, can zoom into different helixes, strands, etc. Good for analyzing amino acid sequences and composition.
Part 4
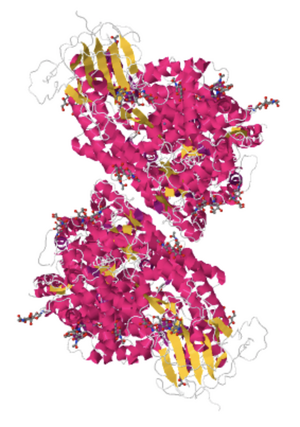
- Selected image from Yan et al. (2020) 6M17
- Interacted with structure and changed it to "JSMol" (JavaScript) format.
- Resulting Structure:
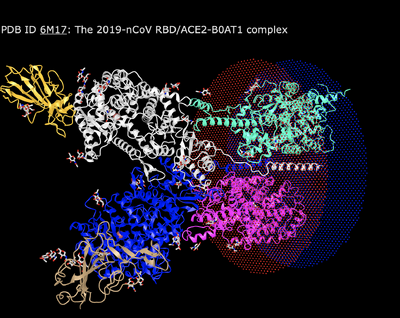
- Used alternate database NCBI Structure home page to look at structure.
- Typed "6M17" into search field, and got the 2019-nCoV RBD/ACE2-B0AT1 complex.
- Selected "full-featured 3D viewer" to view entire 3-D structure.
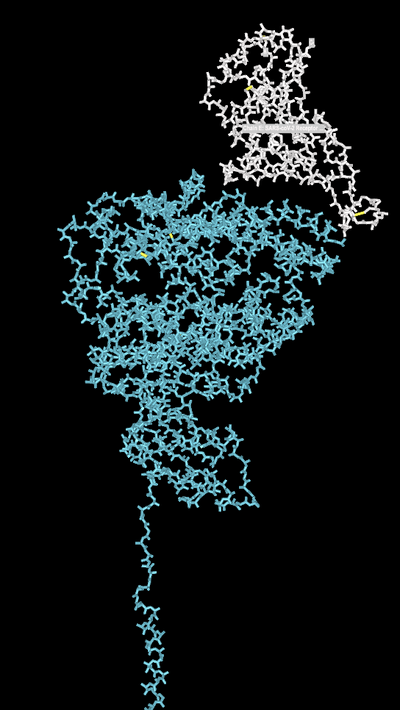
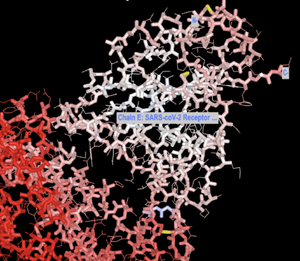
- Resulting image:
- Orienting picture similar to Yan et al.
- Structure from Yan et al.:
- In order to recreate the image from Yan et al.,
- Select the Angiotensin-converting enzyme 2 (Gene: ACE2) and the SARS-coV-2 Receptor Binding Domain
- Click "View" > "View only selection" tabs.
- I then went to "Color" and changed the colors of the strands to similar colors Yan et al. chose.
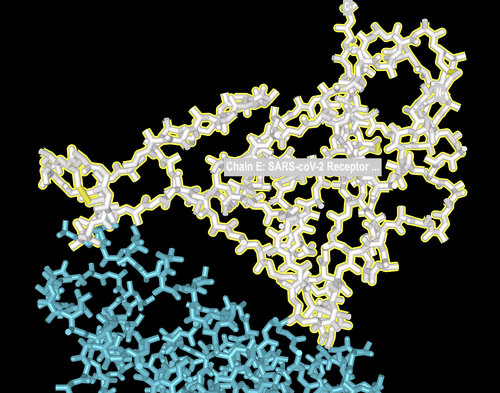
- Resulting Image:
- Image zoomed in:
- Looked at structure on 6M17 once again, and changed format.
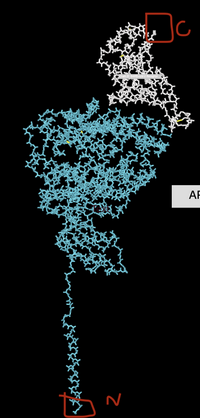
- Took screenshot and drew around N and C terminus'
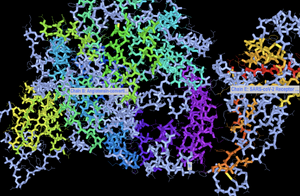
- Secondary Structures were changed to different colors.
- Image with secondary structures:
- The secondary structures of the image are similar to the results seen on Predict Protein.
- The amino acids discussed in the Yan et al. paper are those highlighted in light pink in the following image:
Data and Files
- https://openwetware.org/wiki/File:ProteinstructureKV.png
- https://openwetware.org/wiki/File:FASTAsequenceKV.png
- https://openwetware.org/wiki/File:PredictproteinKV.png
- https://openwetware.org/wiki/File:JavaScriptKV.png
- https://openwetware.org/wiki/File:Full3DstructureKV.png
- https://openwetware.org/wiki/File:Yan_et_al._(2020)_Figure_4A.png
- https://openwetware.org/wiki/File:Fullview.png
- https://openwetware.org/wiki/File:ZoomACE2kv.png
- https://openwetware.org/wiki/File:CandNKV.png
- https://openwetware.org/wiki/File:AllsecondarystructuresKV.png
- https://openwetware.org/wiki/File:AMINOACIDSKV.png
Scientific Conclusion
- The assignment was important in order to grasp an understanding of how the SARS-CoV-02 binds to the human receptor (ACE2). ExPASY Translate tool was first used in order to translate the DNA sequence into a protein sequence. UniProt Knowledgebase (UniProt KB) had information about the spike protein in SARS-CoV-02, that was useful for the experiment. The PredictProtein server server took a while to generate results, but in all, it was important for understanding the spike protein. Finally, the NCBI Structure home page database was the most important for analyzing the protein and structures. The purpose was to get familiar with the tools and be able to highlight which parts of the structure are most crucial. After several attempts, it was easier to become accustomed to how the database works. The task was to recreate the image of the Virus and the ACE2 that was provided in the previous paper by Yan et al.
Research Project Proposal
- What question will you answer about sequence-->structure-->function relationships in a SARS-CoV-2 protein?
- We will be answering how the SARS-CoV-2 protein reacts/binds to ACE2 receptors in various species.
- Species included will be different from those in the Wan 2020 paper.
- What sequences will you use? I want you to take advantage of sequence data available to perform a multiple sequence alignment as part of your project.
- Sequences of the ACE2 receptor.
- Sequencing the ACE2 receptor of species:
- Elephant
- Human
- Mouse
- Rat
- Civet
- Bovine
- Orangutan
- Horse
- Dog
- Platypus
- Chicken
- Opossum
- Chameleon
- Zebrafish
- Bat
- Chinese Softshell Turtle
- Sperm Whale
- Red Fox
- Giant Panda
- Pig
- What protein tools will you use for analysis and answering your question?
- Uniprot will be used in order to collect sequences of species.
- Phylogeny.fr will be used to make various phylogenetic trees relating the species.
- PredictProtein server will be used to confirm information about sequences.
- NCBI Structure home page will be used to look at structures.
Acknowledgements
- I used a photo of Yan et al. structure, that was uploaded by Jack
- I copied the instructions from Week 13
- I texted my group partners Madeleine and Maya to discuss our final research proposal.
- Except for what is noted above, this individual journal entry was completed by me and not copied from another source.
(Kvescio (talk) 22:47, 27 April 2020 (PDT))
References
- OpenWetWare. (2020). BIOL368/S20:Week 13. Retrieved April 20, 2020, from https://openwetware.org/wiki/BIOL368/S20:Week_13
- PredictProtein (2020). RostLab. Retrieved April 26, 2020, from https://open.predictprotein.org/
- UniProtKB - P59594 (2020). National Center for Biotechnology Information (NCBI). Retrieved April 26, 2020, from https://www.uniprot.org/uniprot/P59594
- ExPASy - Translate tool (2020). SIB Swiss Institute of Bioinformatics. Retrieved April 22, 2020, from https://web.expasy.org/translate/
- NCBI. (2020). 6M17: The 2019-nCoV RBD/ACE2-B0AT1 complex. Retrieved April 22, 2020, from https://www.ncbi.nlm.nih.gov/Structure/pdb/6M17.
- Yan, R., Zhang, Y., Li, Y., Xia, L., Guo, Y., & Zhou, Q. (2020). Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science, 367(6485), 1444-1448. doi: 10.1126/science.abb2762.