Angela C Abarquez Week 6
Electronic Lab Notebook
Purpose
By using the data provided by the Microsoft Access Database, an Excel workbook was created for use in GRNmap. This will allow further analyzation of the gene model being developed.
Methods
Creating the GRNmap Input Workbook
The input Excel workbook to be run in the GRNmap modeling software was generated using the following steps. The sample workbook provided by Dr. Dahlquist and Dr. Fitzpatrick was used for guidance (see Data and Files Section).
production_rates sheet
- A new Excel workbook was created. In the first worksheet, labeled "production_rates", the first column was labeled "id" and the second "production_rate".
- The "StandardName" was used in the "id"column. The 11 gene names were copied and pasted from Week 5's 'network' worksheet (used for GRNsight) and pasted under this column header.
- Under the "production_rate" column the initial guesses from the P parameter, rounded to four decimal places, for each corresponding gene was input. The production rates were found in the corresponding table in a Microsoft Access database provided by Dr. Dahlquist and Dr. Fitzpatrick (see Data and Files Section).
degradation_rates sheet
- A second worksheet was added and labeled "degradation_rates".
- The first column was titled "id", and the same gene names were copied and pasted from column 1 of the "production_rates" sheet to this column.
- The next column to the right was labeled "degradation_rate". The data points were copied and pasted (rounded to 4 decimal places) for each corresponding gene from the 'degration_rates' table in the Microsoft Access Database.
Expression Data Sheets for Individual Yeast Strains
- Four more worksheets were added and labeled "wt_log2_expression", "dgln3_log2_expression", "dhap4_log2_expression", and "dzap1_log2_expression".
- The expression data from the wild type and dGLN3, dHAP4, and dZAP1 deletion strains (found in the Microsoft Access Database) was used.
- In each worksheet, the first column was the same copied-and-pasted "id" column from the previous two worksheets (in the same order).
- The next series of columns contain the expression data for each gene at a given timepoint given as log2 ratios (log2 fold changes). The column header was labeled with the time at which the data were collected, without any units. For example, the 15 minute timepoint was labeled "15" and the 30 minute timepoint was labeled "30". Replicate data for the same timepoint was put in columns immediately next to each other and have the same column headers. For example, three replicates of the 15 minute timepoint have "15", "15", "15" as the column headers.
- Data for the 15, 30, and 60 minute timepoints, but not the 90 or 120 minute timepoints was included.
- Each data point was found and then copied and pasted into the appropriate spot.
network sheet
- The network derived from the YEASTRACT database for the Week 5 assignment was copied and pasted into this sheet directly.
- The upper-left cell (A1) was re-labeled “cols regulators/rows targets”.
network_weights sheet
These are the initial guesses for the estimation of the weight parameters, w.
- The "network" sheet was copied and pasted into this worksheet.
optimization_parameters sheet
- This worksheet was copied and pasted from the sample workbook provided (see Data and Files Section).
threshold_b sheet
These are the initial guesses for the estimation of the threshold_b parameters.
- Two columns were created. The first was the same "id" column used previously which was copied and pasted. The second was given the header "threshold_b", and "0" was input for every row for each gene under this column.
Dynamical Systems Modeling of the Gene Regulatory Network
GRNmap (Gene Regulatory Network Modeling and Parameter Estimation) was used to run the model and analyze the results. It is written in MATLAB, therefore the following was performed on a computer in the classroom during class on 2/28.
- The GRNmap v1.10 code was downloaded from the GRNmap Downloads page.
- The file was unzipped by right-clicking -> 7-zip -> Extract here.
- MATLAB R2014b was launched.
- GRNmodel.m was opened, which was in the directory that was unzipped GRNmap-1.10 > matlab.
- The Run button (green "play" arrow) was clicked.
- The program prompted selection of the input workbook.
- An optimization diagnostics graphic that shows the progress of the estimation was shown. When the run finished, expression plots were displayed.
- The output .xlsx and .mat files were saved into a folder labeled "GRNsight and MATLAB results" (found in class Box).
- The output .xlsx file was uploaded onto GRNsight to visualize results.
Results
An Excel workbook (found in the Data and Files Section) was created for use in GRNmap. This will allow for an in-depth analyzation of the gene regulatory networks found in the microarray data from Dr. Dahlquist's cold shock experiment.
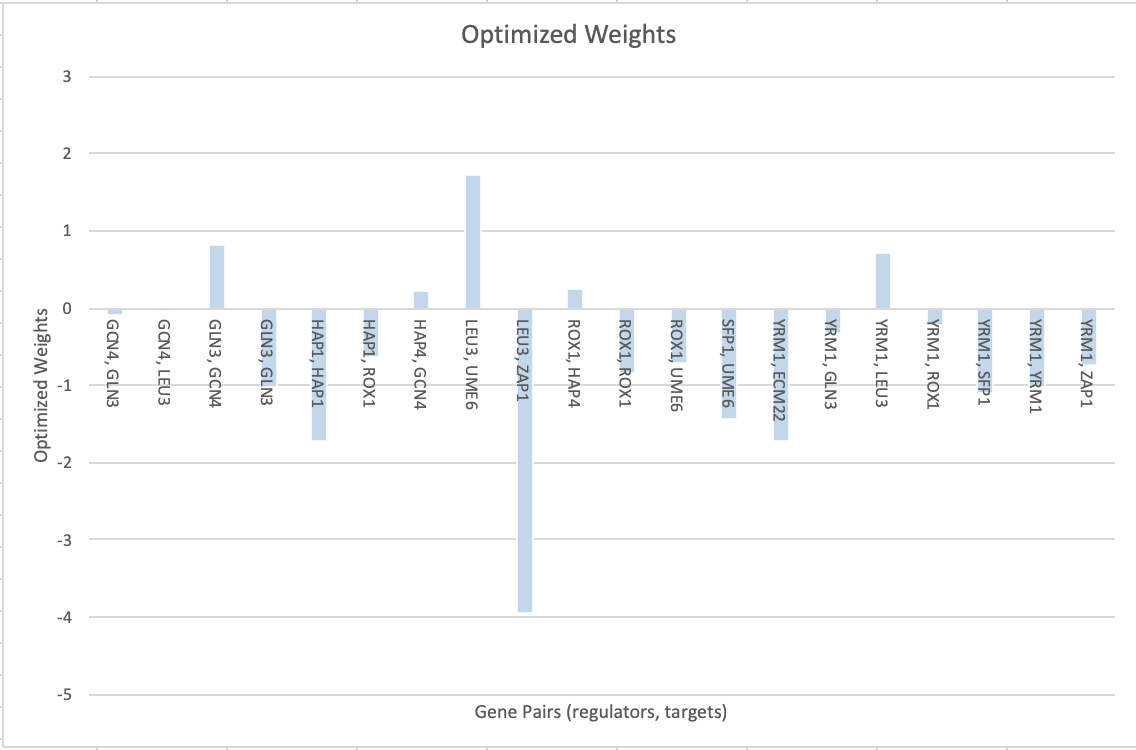
Figure 1. Optimized weights of gene pairs from the output file developed by the MATLAB run.
Data and Files
GRNmap Input Workbook: linked here
Sample workbook: download here
Microsoft Access database: download here
Conclusion
As a result of the work completed in Week 6, an Excel workbook was created to be used in GRNmap. Next steps involve using this software to run the model and analyze the results in an attempt to understand the gene regulatory networks taking place.
Acknowledgments
I worked with my homework partner, Sahil. We texted a few times to clarify details of the assignment.
I also used Dr. Dahlquist and Dr. Fitzpatrick's help in class when we started the assignment.
Except for what is noted above, this individual journal entry was completed by me and not copied from another source. Angela C Abarquez (talk) 19:07, 27 February 2019 (PST)
References
The Week 6 Assignment was used to create this journal entry. Specifically, the "Creating the GRNmap Input Workbook" section was used to describe the methods.
The sample Excel workbook (provided in the Data and Files Section) was referenced when creating the Excel workbook for GRNmap.
Data was retrieved from the Microsoft Access Database provided (see Data and Files Section).
Quick Links
User Page: Angela Abarquez
Course Homepage:BIOL388/S19
Assignments:
Week 1: Instructions and Class Journal
Week 2: Instructions and Class Journal and Individual Journal
Week 3: Instructions and Class Journal and Individual Journal
Week 4/5: Instructions and Class Journal and Individual Journal
Week 6: Instructions and Class Journal and Individual Journal
Week 7/8: Instructions and Class Journal and Individual Journal
Week 9: Instructions and Class Journal and Individual Journal
Week 10: Instructions and Class Journal and Individual Journal
Week 11: Instructions and Class Journal and Individual Journal
Week 12: Instructions and Class Journal and Individual Journal
Week 14/15: Instructions and Individual Journal