AVM BIOL368 Week 6
Purpose
- To determine whether the virus copy numbers of the subjects from the Markham et al. paper are correlated with diversity and divergence.
- To keep track of our research in an electronic notebook and create a powerpoint presentation.
- To use biology workbench to illustrate our findings.
Methods and Results
- Determine final subjects to use for research
- Regroup subjects based on virus copy number
- Classify subjects based on virus copy number
- Calculate p-value
- Calculate theta and S value of subject groups
- Expect to see significantly different values
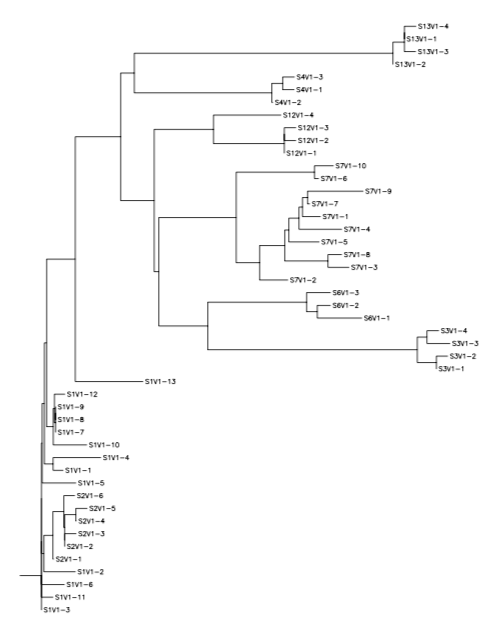
- Create a phylogenetic tree with the last visit from each subject to show which subjects are more closely related
- Use first visit from each subject (will have more diversity) to show relationship
- Use ALL mutations from each visit
- Use first visit from each subject (will have more diversity) to show relationship
- Calculate S value for each subject
- Calculate theta
- Follow protocol from week 4
- Create a phylogenetic tree for each group
- Choose "rooted" view
- Create a rooted tree for all subjects together
- Results show large theta and S values
- Theta values almost identical
- Rejects the hypothesis
- Theta values almost identical
- We found no correlation between genetic diversity and viral load
Data and Files
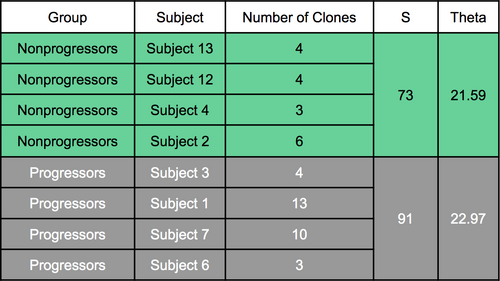
Progressors Grouping for our analysis: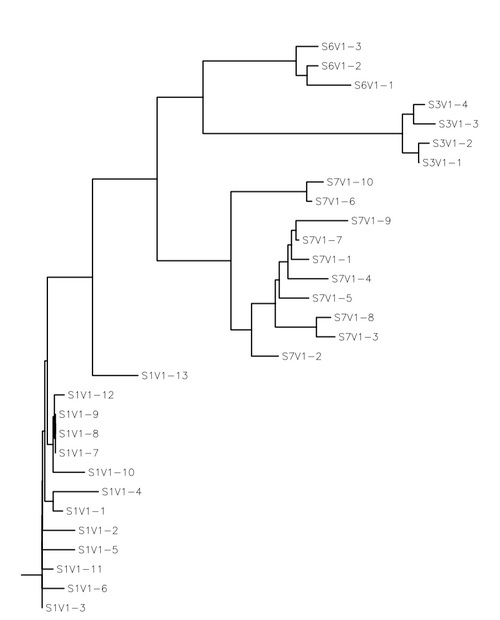
Statistical Data of our grouping of Progressors and Nonprogressors:
Rooted Tree of all subjects:
Conclusion
We used S-value to quantify diversity, theta to estimate genetic distance and generated three rooted phylogenetic trees for the progressors, nonprogressors and all subjects chosen for this study on one tree. Our findings refuted our hypothesis that viral load number would be correlated to genetic diversity. We chose subjects which had high virus copy numbers and subjects with low virus copy numbers. The mutations from the first visit of each subject were used as those were the subjects that the virus copy numbers were calculated from in the Markham et al. paper. The classifcation of subjects based on virus copy number rather than CD4 T-cell count showed no significance and was proved by our findings. The theta values were almost identical for both groups and were both relatively high numbers. That meant that the subjects themselves were not genetically similar to each other and that there was not a significant difference between the subjects classified as progressors and the subjects classified as nonprogressors. The s-values calculated were also very high, indicating that there was a large number of differences between the sequences of each subjects mutations. The subjects were grouped based on subject number on all three of the rooted trees. Each subjects mutations stayed grouped and there was no significance between the subjects who had closer distances on the trees. In conclusion, there is no correlation between virus copy number and diversity.
Acknowledgments
- Worked in collaboration with research partner Courtney L. Merriam in class.
- We also created a shared powerpoint together.
- Template followed from hard-copy protocol as well as our week 6 assignment BIOL368/F16
- Help from our professor, Dr. Dahlquist in class.
- Note: While I worked with the people noted above, this individual journal entry was completed by me and not copied from another source.
Avery Vernon-Moore 21:20, 4 October 2016 (EDT)
References
- Markham, R.B., Wang, W.C., Weisstein, A.E., Wang, Z., Munoz, A., Templeton, A., Margolick, J., Vlahov, D., Quinn, T., Farzadegan, H., & Yu, X.F. (1998). Patterns of HIV-1 evolution in individuals with differing rates of CD4 T cell decline. Proc Natl Acad Sci U S A. 95, 12568-12573. doi: 10.1073/pnas.95.21.12568
- Vlahov, D., Anthony, J.C., Munoz, A., Margolick, J., Nelson, K.E., Celentano, D.D., Solomon, L., Polk, B.F. (1991). The ALIVE study, a longitudinal study of HIV-1 infection in intravenous drug users: description of methods and characteristics of participants. NIDA Res Monogr 109, 75-100.
Weekly Assignments:
Assignment 1 Assignment 2 Assignment 3 Assignment 4 Assignment 5 Assignment 6 Assignment 7 Assignment 8 Assignment 9 Assignment 10 Assignment 11 Assignment 12 Assignment 14 Assignment 15
Individual Journals:
"Week 1: Create Wiki Page" Individual Journal 2 Individual Journal 3 Individual Journal 4 Individual Journal 5 Individual Journal 6 Individual Journal 7 Individual Journal 8 Individual Journal 9 Individual Journal 10 Individual Journal 11 Individual Journal 12 No Week 13 Assignment Individual Journal 14 Individual Journal 15
Shared Journals:
Class Journal 1 Class Journal 2 Class Journal 3 Class Journal 4 Class Journal 5 Class Journal 6 Class Journal 7 Class Journal 8 Class Journal 9 Class Journal 10 Class Journal 11 Class Journal 12 Class Journal 14 Class Journal 15