AVM BIOL368 Week 4
Purpose
- To continue with our class research and journal projects for the Markham et al. paper
- Continue to learn about sequencing and the similarities and differences
Methods and Results
Part 1
- Upload the “visit_1_S1_S9.txt” and “visit_1_S10_S15.txt” files from the BIOL368/F16 Week 4 page into the Biology Workbench
- Generate a multiple sequence alignment using ClustlW
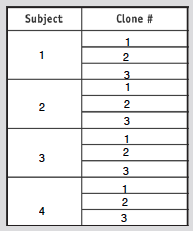
- Choose 3 clones form 4 different subjects (as shown in table below)
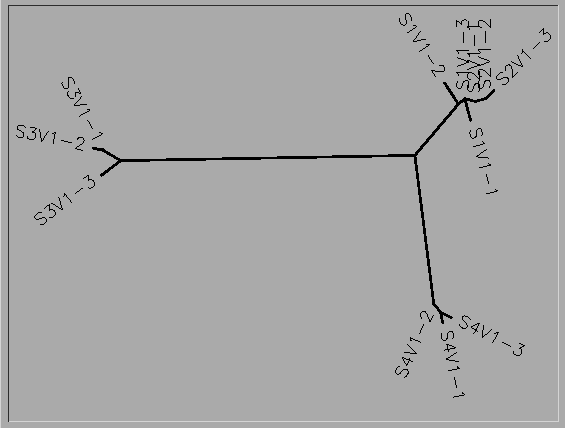
- Photo of tree shown below
Questions
- Do the clones from each subject cluster together?
- Yes
- Do some subjects’ clones show more diversity than others?
- Subject 1, strain 1 is more diverse than strains 2 and 3. Subject 2 strain 3 is more diverse than stains 1 and 2. Subject 3 strain 3 is more diverse than strains 1 and 2. Subject 4 strain 2 is more diverse than strains 1 and 3.
- Do some of the subjects cluster together?
- Yes. Subjects 1 and 2 are clustered.
- Write a brief description of the tree and how to interpret the clustering pattern with respect to the similarities and potential evolutionary relationships between subjects’ HIV sequences.
- The unrooted tree shows the similarities of the 4 subjects and clones. This tree shows that Subject 1 and 2 have very closely related viruses, and within this virus, mutations 1 and 2 of subject 2 are most closely related to mutation 3 of subject 1. All of the mutations of subject 4's virus are most closely related to each other and the mutations of subject 3 are most closely related to each other.
Part 2
- Select a sequence from last weeks assignment and align all of the clones
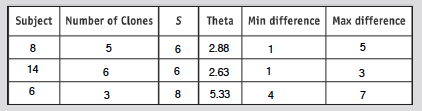
- Calculate S from the alignment
- S is the number of positions where there is at least one nucleotide difference across the clones
- Save the alignment for later use
- Run an analysis for two more subjects
- Sequence differences for each subject shown at right



- (Math Is Fun was used to calculate theta)
- Within the Clustdist tool generate a distance matrix for one of your alignments
- Choose the highest and lowest pairwise scores and find the % difference
- Calculate theta for each subject
- Repeat these steps for each subject
- Table below shows results from these analysis
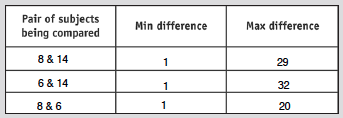
- Create a new alignment with all the sequences from 2 different subjects
- Use Clustdist to create a pairwise distance matrix
- Find all the min and max differences
- Results shown in table below
Data and Files
- All files were saved to an external hard drive as well as on this page
- HIV Sequences, Visit 1, Subjects 1-9
- HIV Sequences, Visit 1, Subjects 10-15
Conclusion
This exercise was done to determine if the HIV virus strains extracted from random test subjects originated from the same virus initially.Clones from subject 3 are the most genetically different of the subjects used. The tree shows that subjects 1 and 2 had very similar viruses, with some strains (subject 2; mutations 1 and 2, and subject 1; mutation 3) were even mores genetically similar. Subject 4 was slightly separated as well, but not as far away on the unrooted tree as subject 3. After following the protocol above and creating the unrooted tree during the exercise I understood that the extracted viruses did not stem from one original viral source. Overall, this project was helpful in helping to understand the significance of S values, theta, ClustalWdist, min and max differences and generally knowing how to understand similarities and differences between the sequences given.
Research Project
- What is your question?
- If HIV-1 virus evolution patterns are based on virus copy number rather than CD4 T-cell decline, how would this change the subjects classification as non-progressors, moderate progressors, and rapid progress ors? Is virus copy number correlated with diversity and divergence?
- Make a prediction (hypothesis) about the answer to your question before you begin your analysis.
- There would be a shift in the subjects being grouped as moderate and rapid progressors. There is an overlap in virus number copies in Table 1 for subjects in both of these groups.
- Which subjects, visits, and clones will you use to answer your question? Justify why you chose the subjects, visits, and clones you did.
- We could look at subjects 1, 3, 6 and 7 as they have very similar virus number copies. We could also choose subjects 12 and 13 to compare to those which have significantly lower virus number copies. For this specific question I think almost any of the subjects could be used and it may need to be adjusted once we actually start to work on the project.
Acknowledgments
- Worked in collaboration with Courtney L. Merriam in class. Courtney and I agreed on the topic and details of our research in class, and wrote the section titled "Defining Our Research Project" together.
- Template followed from week 4 assignment BIOL368/F16
- Help from our professor, Dr. Dahlquist in class.
- Note: While I worked with the people noted above, this individual journal entry was completed by me and not copied from another source.
Avery Vernon-Moore 14:42, 21 September 2016 (EDT)
References
- Donovan S and Weisstein AE (2003) Exploring HIV Evolution: An Opportunity for Research. In Jungck JR, Fass MR, and Stanley ED, eds. Microbes Count! West Chester, Pennsylvania: Keystone Digital Press.
- Markham, R.B., Wang, W.C., Weisstein, A.E., Wang, Z., Munoz, A., Templeton, A., Margolick, J., Vlahov, D., Quinn, T., Farzadegan, H., & Yu, X.F. (1998). Patterns of HIV-1 evolution in individuals with differing rates of CD4 T cell decline. Proc Natl Acad Sci U S A. 95, 12568-12573. doi: 10.1073/pnas.95.21.12568
- Vlahov, D., Anthony, J.C., Munoz, A., Margolick, J., Nelson, K.E., Celentano, D.D., Solomon, L., Polk, B.F. (1991). The ALIVE study, a longitudinal study of HIV-1 infection in intravenous drug users: description of methods and characteristics of participants. NIDA Res Monogr 109, 75-100.
- Week 4 Assignment Page
Weekly Assignments:
Assignment 1 Assignment 2 Assignment 3 Assignment 4 Assignment 5 Assignment 6 Assignment 7 Assignment 8 Assignment 9 Assignment 10 Assignment 11 Assignment 12 Assignment 14 Assignment 15
Individual Journals:
"Week 1: Create Wiki Page" Individual Journal 2 Individual Journal 3 Individual Journal 4 Individual Journal 5 Individual Journal 6 Individual Journal 7 Individual Journal 8 Individual Journal 9 Individual Journal 10 Individual Journal 11 Individual Journal 12 No Week 13 Assignment Individual Journal 14 Individual Journal 15
Shared Journals:
Class Journal 1 Class Journal 2 Class Journal 3 Class Journal 4 Class Journal 5 Class Journal 6 Class Journal 7 Class Journal 8 Class Journal 9 Class Journal 10 Class Journal 11 Class Journal 12 Class Journal 14 Class Journal 15