Taylor Makela Journal Week 5
From OpenWetWare
Jump to navigationJump to search
Taylor Makela Week 5
Purpose
- The purpose of this assignment was to use the web-based iCn3D viewer to explore the spike protein structure of SARS-CoV-2, to do our own analysis of Figure 1C SARS-CoV RBD (optimized for human ACE2 recognition) and human ACE2: 3SCI, and to do an analysis of civet ACE2-spike protein structure shown in Figure 4B. Lastly, this assignment provided as a basis to begin thinking about our research project.
Methods and Results
Exploring the Spike Protein Structure
- Navigated to UniProt Knowledgebase (UniProt KB) to discover what is already known about the spike protein for SARS-CoV-2.
- Searched for the keyword "SARS-CoV-2" in the main UniProt search field.
- Observed 1,616 results in the main UniProt search field.
- Some of the results are viral proteins and some of the results are methyltransferases.
- Navigated to the entry for the SARS-CoV-2 spike protein entry with accession number "P0DTC2".
- In this protein data base entry, there is information regarding the function, location, taxonomy, pathology, molecule processing, amino acid modifications, interactions, domains, and sequencing of the SARS-CoV-2 spike protein.
- Used the web-based iCn3D viewer to look at the different SARS-CoV structures provided, and decided to select and focus on Figure 1C SARS-CoV RBD (optimized for human ACE2 recognition) and human ACE2 (optimized for human ACE2 recognition) and human ACE2: 3SCI
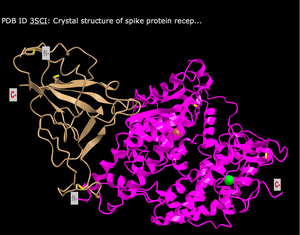
Recreating Figure 1A from the Wan et al. (2020) Paper
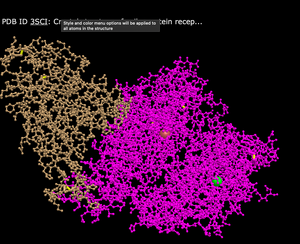
- Image of Figure 1A recreation:
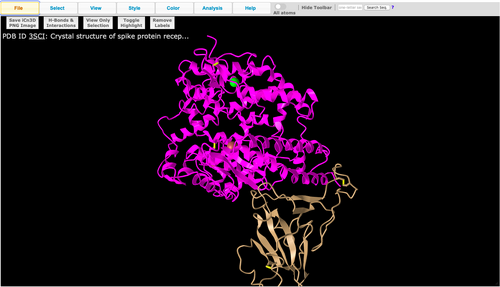
- Is this a primary, secondary, tertiary, or quaternary structure? Explain your reasoning.
- This is a quaternary structure because it includes two polypeptide chains that interact together (the ionic bond between E35 on ACE2 and R479, and the H-bond between T31 on ACE2 and Y442).
- How many domains are in this structure? Explain your reasoning.
- There are two domains in this structure - represented by the pink (the ACE2 protein) and tan (spike protein) colors shown in the image above.
Various Protein Styles
- To look at various protein styles, I navigated to the "Style" tab at the top of the page, and then clicked on the following protein styles:
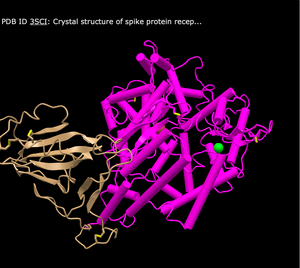
- Cylinder and Plate
- What's Unique: Has distinct lines and cylinders to depict the structure
- Cylinder and Plate
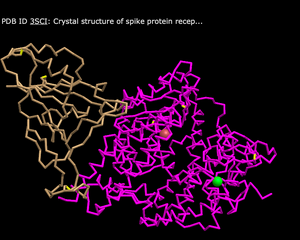
- C Alpha Trace
- What's Unique: Has thin lines and curls to depict the structure
- C Alpha Trace
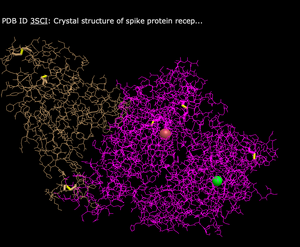
- Lines
- What's Unique: Has thin lines with branches to depict the structure
- Lines
- Ball and Stick
- What's Unique: Has balls to represent different compounds and lines to represent various bonds in the structure
- Ball and Stick
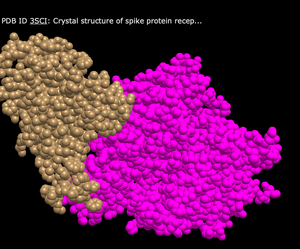
- Spheres
- What's Unique: Uses closely condensed spheres that show the overall shape of the structure
- Spheres
Various Color Schemes in the "Spheres" View
- To look at various color schemes, I navigated to the "Color" tab at the top of the page, and then clicked on the following color schemes:
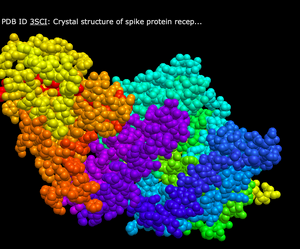
- Spectrum
- What's Unique: Shows very bright and distinct colors that are separated in various regions throughout the structure
- Spectrum
- Secondary
- What's Unique: This color scheme has a yellow sheet, and either blue or red for the various proteins within the structure
- Secondary
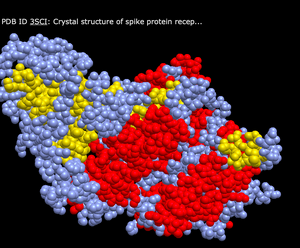
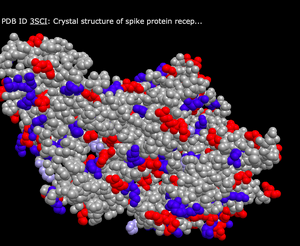
- Charge
- What's Unique: This color scheme shows either grey, red, blue, or light purple colors to depict the various proteins within the structure
- Charge
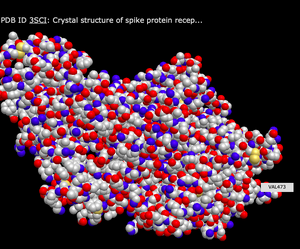
- Atom
- What's Unique: This color scheme shows either blue, grey, red, or yellow depending on the atom
- Atom
N-terminus and C-terminus of Each Polypeptide
- To view the N-terminus and C-terminus of each polypeptide, I first navigated to the "Analysis" tab at the top of the page, found the "Label" section, and then selected "N- & C- Termini" option.
Secondary Structures Found in the ACE2 Protein
- The ACE2 protein (shown in magenta) consist of mostly of alpha helices (the ribbon like rings), but also contains beta sheets (the thicker lines with thin loops at the end).
Secondary Structures Found in the Spike Protein
- The Spike protein (shown in gold) consists mainly of beta sheets, which are represented by the thicker lines with thin loops at the end.
Side Chain-Side Chain Interactions in the Civet ACE2-Spike Protein Structure
- Clicked on the link to interact with the structure in iCn3D.
- Clicked on the Windows menu to “View Sequences & Annotations”.
- In the new window that appeared to the right, I clicked on the “Details” tab to show the actual amino acid sequences.
- There were 2 sets of ACE2-spike proteins because of the way the proteins crystallized.
- Focused on the pink and tan chains and oriented them like shown in Figure 4B.
- We are going to be making the amino acid side chains shown in the figure visible.
- In the sequence window, I went to sequence “Protein 3SCK_A” (in pink) and selected the following amino acids
- T31
- E35
- E38
- T82
- K353
- The part of the ribbon that represents these amino acids should be highlighted in yellow in the structure
- Went to the Styles menu and selected Proteins > Ball and Stick
- Went to the Color menu and selected Atom
- Saw the side chains shown in the figure
- Repeated this process for the tan spike protein sequence 3SCK_E for the following amino acids:
- T487
- R479
- G480
- Y442
- P472
- Screenshot oriented like in Figure 4B:
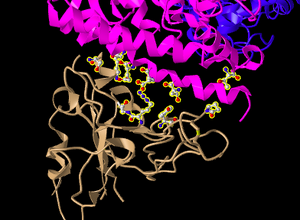
The Ionic Bond Between E35 on ACE2 with R479 on Spike Protein
- E35 on ACE2 makes an ionic bond with R479 on spike protein. Which of these is acidic and which is basic?
- E35 has a positive charge and is therefore acidic, while R479 has a negative charge and is therefore basic.
- How can you tell that from the image you created?
- This is seen by the colors of the atoms in the image. The oxygen of the E35 and the nitrogen of the R479 can be seen, and thus the charges of the amino acids can be inferred.
The Hydrogen Bond Between T31 on ACE2 with Y442 on Spike Protein
- T31 on ACE2 makes a hydrogen bond with Y442 on spike protein. Which side chain classification do these two amino acids belong to (acidic, basic, uncharged polar, nonpolar)?
- Both T31 and Y442 have OH groups, making them basic. Both T31 and Y442 are uncharged polar.
- How can you tell that from the image you created?
- This can be seen in the image because the oxygens of the OH groups in both amino acids are visible (blue) thus indicating that that these amino acids are basic and uncharged polar.
3D Display Showing Dashed Lines Representing the Ionic Bonds and H-bonds Between the 2 Polypeptide Chains and Amino Acids
- Went to the View menu and selected H bonds & Interactions
- In part 1 of the window that appeared, I unchecked “Contacts/Interactions” leaving Hydrogen Bonds and Ionic Interaction checked
- In part 2 of the window, I selected the first set “3SCK_A” (pink)
- In part 3 of the window, I selected the second set “3SCK_E” (tan)
- In part 4 of the window, I clicked on the button “3D Display”
- I then saw the dashed lines representing the ionic bonds and H-bonds between the two polypeptide chains and amino acids we described above:
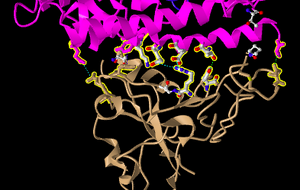
Beginning My Research Project
- Answering questions with my homework partner, Nida Patel:
- What question will you answer about sequence-->structure-->function relationships in the spike and/or ACE2 protein?
- Is COVID-19 transmission from a human host to 14 other species possible based on their individual ACE2 sequences?
- What sequences will you use?
- Human: ACE2_HUMAN Angiotensin-converting enzyme 2
- Mouse: ACE2_MOUSE Angiotensin-converting enzyme 2
- Rat: ACE2_RAT Angiotensin-converting enzyme 2
- Civet: CE2_PAGLA Angiotensin-converting enzyme 2
- Chinese Rufous Horseshoe bat: AGZ48803.1 angiotensin-converting enzyme 2 [Rhinolophus sinicus]
- Pig: tr|K7GLM4|K7GLM4_PIG Angiotensin-converting enzyme
- Chinese Pangolin: QLH93383.1 angiotensin I converting enzyme 2 [Manis pentadactyla]
- Ferret: sp|Q5RFN1.1|ACE2_PONAB Angiotensin-converting enzyme 2
- Cat: sp|Q5RFN1.1|ACE2_PONAB Angiotensin-converting enzyme 2
- Dog: sp|Q5RFN1.1|ACE2_PONAB Angiotensin-converting enzyme 2
- Monkey: AAY57872.1 angiotensin converting enzyme 2
- Orangutan: sp|Q5RFN1.1|ACE2_PONAB Angiotensin-converting enzyme 2
- Will need to work on making a multiple sequence alignment, phylogenetic tree, and protein structures.
Data and Files
- UniProt
- NCBI Structure Database
- MN908947: Severe acute respiratory syndrome coronavirus 2 isolate Wuhan-Hu-1 genome sequence
- 3SCI Spike Protein of SARS-CoV RBD (optimized for human ACE2 recognition) and Human ACE2
- civet ACE2-spike protein structure
Scientific Conclusion
This assignment allowed me to practice using the web-based iCn3D viewer in order to learn about the structures of the spike protein of SARS-CoV-2, Figure 1C SARS-CoV RBD (optimized for human ACE2 recognition) and human ACE2: 3SCI, and civet ACE2-spike protein structure shown in Figure 4B. This taught me how to manipulate structures to be able to visualize atoms, bonds, and secondary, tertiary, and quaternary structures.
Acknowledgments
- I acknowledge my homework partner Nida Patel, who I consulted for several hours regarding syntax, formatting, and content questions.
- I acknowledge that I copied and modified the format, syntax of Anna Horvath Week 5 Assignment Page
- I acknowledge that I copied and modified the protocol shown on the Week 5 assignment page for this course.
Except for what is noted above, this individual entry was completed by me and not copied from another source. Taylor Makela (talk)
References
- iCn3D: Web-based 3D Structure Viewer 3SCK. (2020). Retrieved 14 October 2020, from https://www.ncbi.nlm.nih.gov/Structure/icn3d/full.html?pdbid=%203SCK
- iCn3D: Web-based 3D Structure Viewer 3SCK. (2020). Retrieved 14 October 2020, from https://www.ncbi.nlm.nih.gov/Structure/icn3d/full.html?pdbid=%203SCK
- OpenWetWare. (2020). BIOL368/F20:Week 5. Retrieved 14 October 2020, from https://openwetware.org/wiki/BIOL368/F20:Week_5
- Uniprot. (2020). S - Spike glycoprotein precursor - Severe acute respiratory syndrome coronavirus 2 (2019-nCoV) - S gene & protein. Retrieved 14 October 2020, from https://www.uniprot.org/uniprot/P0DTC2
- Wan, Y., Shang, J., Graham, R., Baric, R., & Li, F. (2020). Receptor Recognition by the Novel Coronavirus from Wuhan: an Analysis Based on Decade-Long Structural Studies of SARS Coronavirus. Journal Of Virology, 94(7). doi: 10.1128/jvi.00127-20
Template
Template Links
Assignment Pages
- Week 1 Assignment Page
- Week 2 Assignment Page
- Week 3 Assignment Page
- Week 4 Assignment Page
- Week 5 Assignment Page
- Week 6 Assignment Page
- Week 7 Assignment Page
- Week 8 Assignment Page
- Week 9 Assignment Page
- Week 10 Assignment Page
- Week 11 Assignment Page
- Week 12 Assignment Page
- Week 14 Assignment Page
Individual Journal Pages
- Taylor Makela Journal Week 2
- Taylor Makela Journal Week 3
- Taylor Makela Journal Week 4
- Taylor Makela Journal Week 5
- Taylor Makela Journal Week 6
- Taylor Makela Journal Week 7
- FoldamerDB Review
- Taylor Makela Journal Week 9
- Taylor Makela Journal Week 10
- Taylor Makela Journal Week 11
- Comparison of Human and Hamster ACE2 Receptors for SARS-CoV-2 Week 12
- Comparison of Human and Hamster ACE2 Receptors for SARS-CoV-2 Week 14