Jordan T. Detamore Week 6
Purpose
The purpose of this process was to attempt to subtype the different sequences from the Markham et al. study
Methods & Results
- Sequences were randomly chosen from the Markham study and another study by Collinson-Streng et al. where sequences were organized by subtype
- After cutting the Collinson-Streng et al. sequences to match the length of the Markham et al. sequences the two groups of sequences were compared
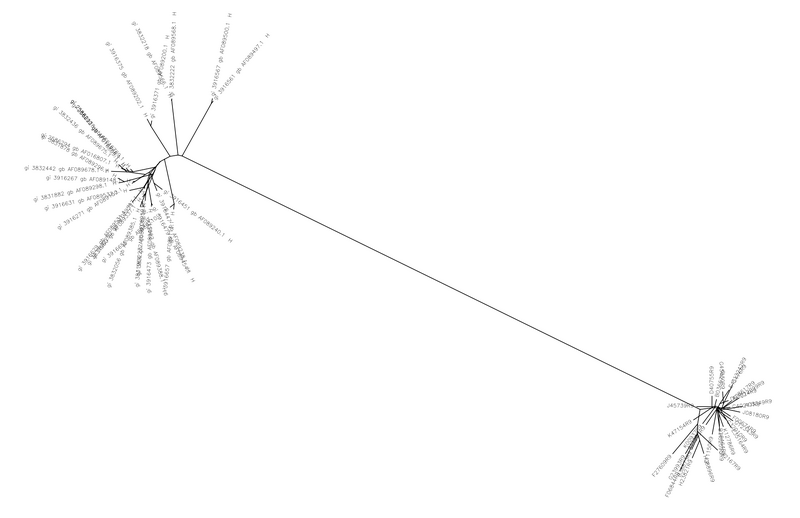
- A ClustalW sequence alignment was used to align the sequences and produce a tree
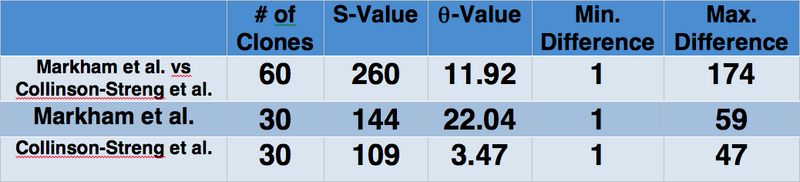
- S-value and theta were computed from the alignment and trees
- Minimum and Maximum difference were calculated using ClustalDist
- The data and trees show that the sequences from Markham et al. were not genetically similar to those from Collinson-Streng et al.
Data & Files
The PDF version of our powerpoint presentation can be found here.
Scientific Conclusion
We were unable to subtype the sequences from the Markham et al. paper. This was due to the fact that the sequences from the Collinson et al. paper were from a study in Uganda in 2009 while the Markham paper studied subjects in Baltimore in 1998. For this reason the subjects from the different studies were very genetically different. We found that the sequences from Markham were more diverse and also that subjects of the same subtype were not always genetically similar. This was our most intriguing, difficult to explain result.
Acknowledgements
- I worked with Matthew K. Oki in class and also used parts of his Electronic Workbook that we share on this weeks assignment
- I received instruction from Dr. Dahlquist's in class as well
- While I worked with the people noted above, this individual journal entry was completed by me and not copied from another source.
- Jordan T. Detamore 23:47, 10 October 2016 (EDT):
References
Collinson-Streng, A. N., Redd, A. D., Sewankambo, N. K., Serwadda, D., Rezapour, M., Lamers, S. L., … Laeyendecker, O. (2009). Geographic HIV Type 1 Subtype Distribution in Rakai District, Uganda. AIDS Research and Human Retroviruses, 25(10), 1045–1048. http://doi.org/10.1089/aid.2009.0127
Markham, R.B., Wang, W.C., Weisstein, A.E., Wang, Z., Munoz, A., Templeton, A., Margolick, J., Vlahov, D., Quinn, T., Farzadegan, H., & Yu, X.F. (1998). Patterns of HIV-1 evolution in individuals with differing rates of CD4 T cell decline. Proc Natl Acad Sci U S A. 95, 12568-12573. doi: 10.1073/pnas.95.21.12568
Vlahov, D., Anthony, J.C., Munoz, A., Margolick, J., Nelson, K.E., Celentano, D.D., Solomon, L., Polk, B.F. (1991). The ALIVE study, a longitudinal study of HIV-1 infection in intravenous drug users: description of methods and characteristics of participants. NIDA Res Monogr 109, 75-100.
Useful links
LMU Seaver College of Science and Engineering