Jordan T. Detamore Week 3
Purpose
The purpose of this assignment was to learn how to use sequences and the biology workbench while starting to work with HIV
Methods & Results
Activity I-Part II
What was the accession number of the sequence you chose?
- AFO89115
Which Subject of the study was that HIV sequence from?
- Subject 3
Which section of the record contains information about who the HIV was collected from?
- The last number of the ID, in this case Clone 3 from S3V3_3
Dowload several sequences in FASTA format
Activity I-Part III
Log into Biology Workbench
Select Nucleic sequence data
Select Add New Sequences and press Run
Select file saved from NCBI
Chose Save to import data into Biology Workbench
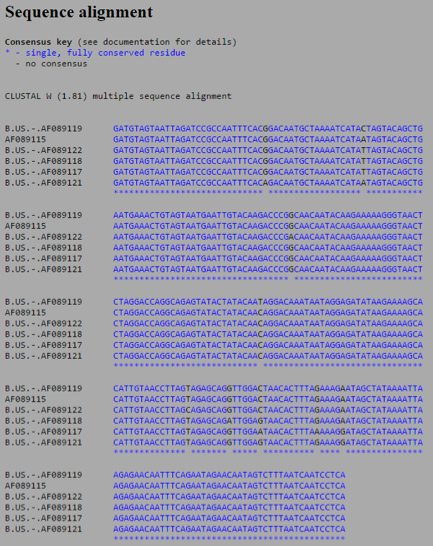
Select all of the sequences and run a multiple sequence alignment using ClustalW
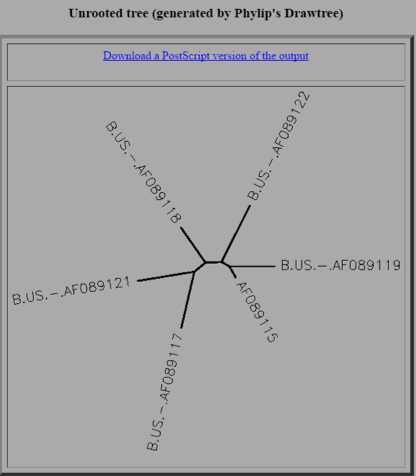
Look over the output and see if you can relate the differences in the sequences to the topology of the unrooted tree diagram and the pairwise similarity scores
Store the sequence
- From both the sequences and the tree you can see that many of the sequences are quite different
- The branches from the tree are very spread apart and it looks as though no two sequences are that similar
- In the sequences you can see that 7 sites where at least one sequence differs if not many
Data & Files
Media:DetamoreJordanFastaHIVS3V33.docx
Scientific Conclusion
Based on the purpose of this assignment which was to learn how to use sequences and Biology Workbench in order to analyze HIV sequences, I was able to successfully complete the assignment. I generated my tree diagram and was able to analyze the differences in the sequences of HIV using the tree and the given sequences
Preparation for Week 4 Journal Club
Definitions
- Seroconversion: The stage in an immune response when antibodies to the infecting agent are first detected in the bloodstream. For example, people infected with HIV typically seroconvert about 4–6 weeks following the initial infection, when antibodies against viral proteins are first produced
- Seropositive: giving a positive result in a test of blood serum, e.g., for the presence of a virus
- Virologic: the study of viruses and the diseases they cause
- CD4 T-Cell:A form of T lymphocyte with CD4 receptor on the cell surface that recognizes antigens of a virus-infected cell.
- Peripheral blood mononuclear cells (PBMC):A peripheral blood mononuclear cell (PBMC) is defined as any blood cell with a round nucleus (i.e. a lymphocyte, a monocyte, or a macrophage). These blood cells are a critical component in the immune system to fight infection and adapt to intruders.
- Epitopes:The structure on the surface of an antigen that is recognized by and can bind to a specific antibody. Its shape is generally complementary to that of the antibody's antigen-binding site.
- Proliferation: An increase in the size of a population of cells.
- Cytolysis: The dissolution or disruption of cells, especially by an external agent.
- Epidemiological: Relating to the branch of medicine which deals with the incidence, distribution, and control of diseases
- Epitopes: the antigenic determinant on an antigen to which the paratope on an antibody binds.
Introduction
- HIV-1 has high mutation and replication rates allowing it to adapt to changes fast in the host environment
- In a "best fit" environment, there would be a dominant mutation and low diversity
- Examining the patterns of diversity can provide information into viral evolution
- Previous studies have used small cohorts of infects subjects, used techniques that did not involve sequence patterns, and have only used a limited number of time points when analyzing each subject
- In this study HIV-1 evolution was studied in 15 subjects at very frequent intervals over four years
- The study found different patterns of selection between nonprogressors and progressors and also that genetic diversity is associated with decline of CD4 T-Cells
Methods
- 15 participants were selected from injection drug users from AIDS Linked to Intravenous Experiences (ALIVE).
- The participants had their blood tested in 6-month intervals for virology and immunologic studies
- Rapid progressors were labeled as those who had less than 200 CD4 T-Cells within 2 years
- Moderate progressors were labeled as those who had between 200 and 650 CD4 T-Cells
- Nonprogressors were labeled as those who had over 650 CD4 T-Cells
- Nested PCR was used to amplify a 285-bp region from peripheral blood mononuclear cells
- First and second round PCR were run
- Reverse transcription PCR was then used to determine the plasma viral load
- Phylogenetic trees were then computed from the sequences
- Seventy-Six time points were taken from the 15 subjects
- The initial consensus sequence was computed for each subject
- The sequences were compared with the observed strain
- When comparing the two strains it was determined whether they were synonymous or non-synonymous
- The inherent bias towards non synonymous changes was corrected by adjusting the number of sites where the mutation could occur
- Three different phylogenetic trees were constructed
- For each individual a regression line of divergence/diversity over time was made
- A random effects model was then used to compare the decline of CD4 T-cell counts
Results
- Patterns of CD4 varied amongst the 15 subjects
- Range:Increase of 53 cells/year-593 cells/year
- Serum viral load data ranged from 1,702 to 321,443 copies of viral genomic RNA/ml
- 873 clones were sequenced and analyzed
- Changes in HIV-1 sequences over time were quantified by two parameters
- Genetic diversity at each visit (-2.94 to 5.10 nt per clone per year)
- Divergence at each visit (0.13% to 2.09%)
- Viruses from initial visits were relatively heterogeneous while later visits were more homogenous
- Diversity and divergence increased over time in all progressor categories
- Diversity and Divergence per year correlated positively with how progressive the subject was
- No strain was proved to be dominant based on phylogenetic trees
Discussion
- HIgher levels of genetic diversity and divergence in HIV-1 variants were found to be associated with a decline in CD4 T-Cells.
- Nonsynonymous substitution rates in progressors were almost three times those in nonprogressors.
- Viral strains showed possible selection against amino acid change in nonprogressors
- Nonprogressors showed selection for amino acid change
- One study by McDonald et al. is in agreement with this study in that genetic diversity increases with rapid progressors
- The study by McDonald et al. observed rapid progressors to have less diversity than slow progressors contrary to this study
- Wolinsky et al. also observed less diversity in the most rapid progressors contrary to this study
- Nowak found similar association between CD4 T- Cell decline and increased diversity and divergence
- While great observations have been made in this study, only 15 subjects were studied and other studies have contradicted this one
- Increasing the sample size could add more strength to this study and continue to find out more about HIV-1
Acknowledgements
- I worked with my lab partner Shivum A Desai in class on this assignment
- I received instructions and guidance on how to complete this assignment from Dr. Dahlquist's Bioinformatics Lab
- While I worked with the people noted above, this individual journal entry was completed by me and not copied from another source.
- Jordan T. Detamore 00:22, 20 September 2016 (EDT):
References
- Week 3 Assignment created by Kam D. Dahlquist
- Subramaniam, S. (1998) The Biology Workbench--a seamless database and analysis environment for the biologist. Proteins, 32, 1-2.
- Donovan S and Weisstein AE (2003) Exploring HIV Evolution: An Opportunity for Research. In Jungck JR, Fass MR, and Stanley ED, eds. Microbes Count! West Chester, Pennsylvania: Keystone Digital Press.
- Markham, R.B., Wang, W.C., Weisstein, A.E., Wang, Z., Munoz, A., Templeton, A., Margolick, J., Vlahov, D., Quinn, T., Farzadegan, H., & Yu, X.F. (1998). Patterns of HIV-1 evolution in individuals with differing rates of CD4 T cell decline. Proc Natl Acad Sci U S A. 95, 12568-12573. doi: 10.1073/pnas.95.21.12568
- Vlahov, D., Anthony, J.C., Munoz, A., Margolick, J., Nelson, K.E., Celentano, D.D., Solomon, L., Polk, B.F. (1991). The ALIVE study, a longitudinal study of HIV-1 infection in intravenous drug users: description of methods and characteristics of participants. NIDA Res Monogr 109, 75-100.
Useful links
LMU Seaver College of Science and Engineering