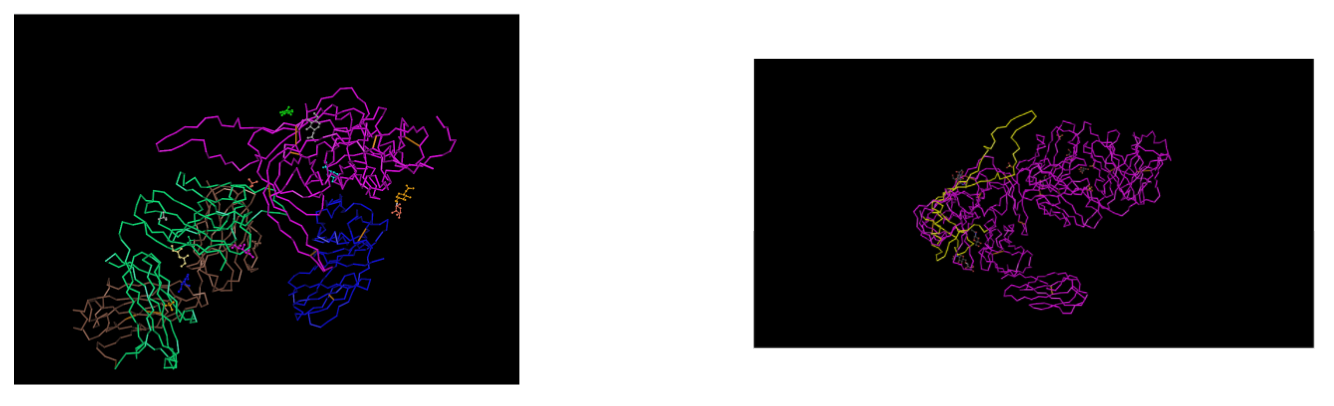Jordan T. Detamore Week 10
Purpose
The purpose of this week was to continue our projects working with the structure and function of HIV in relation to amino acids, while using different programs to work with our given data.
Methods/Results
- Downloaded the 3D sequence of Huang et al. (2005) structure 2B4C gp120 protein using the Cn3D software site.
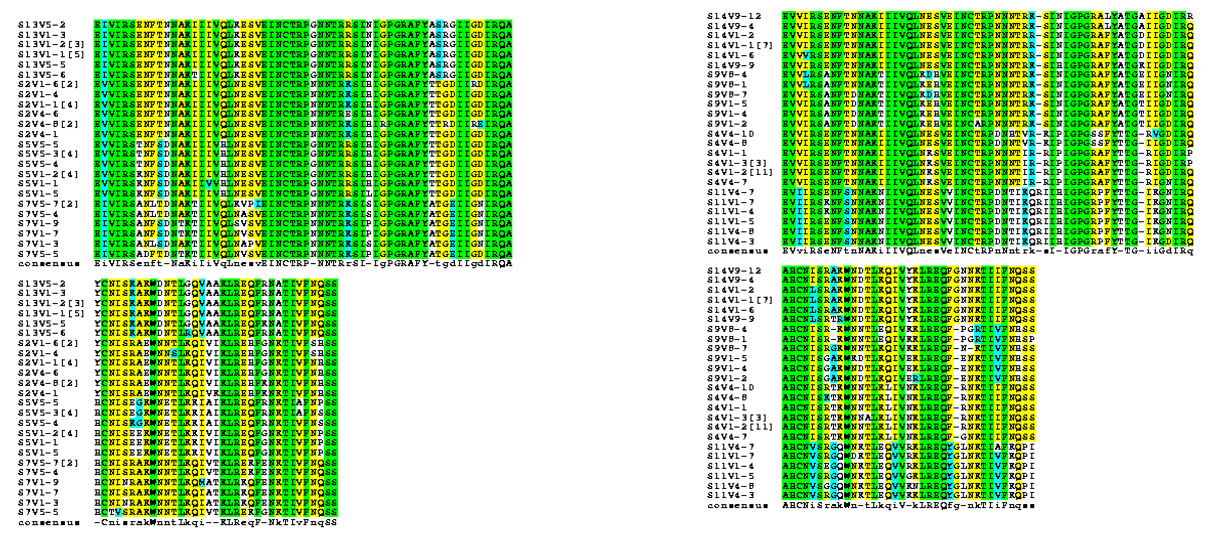
- Using amino acid sequences from the Markham data we identified the region that corresponds with the V3 region of the gp120 protein highlighted in yellow EVV-HSS region.
- We were able to identify the N and C terminals of the tertiary structures by selecting the amino acid and observing which portion of the protein got highlighted.
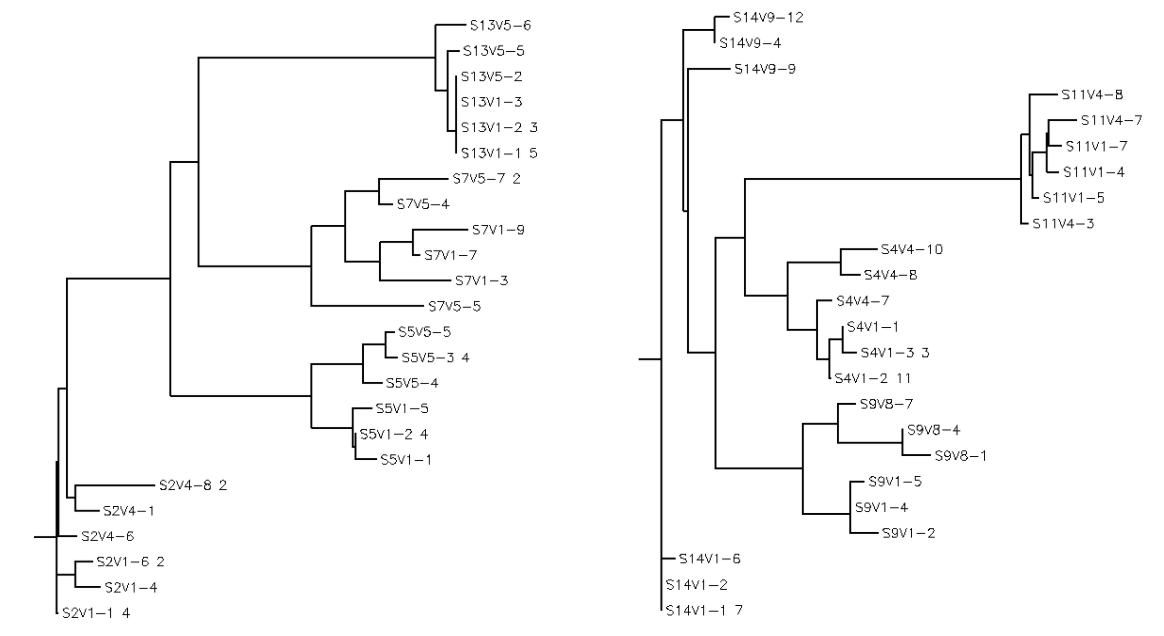
- Amino acid sequences from selected subjects were run under a multiple sequence alignment and a rooted tree was created.
- Strong groups means like for like (acidic for acidic, etc...), weak groups means (hydrophobic for hydrophyllic), non-consensus means there is high variability between amino acids in each sequences. We decided to run two alignments to compare how many of these types of substitutions occur in the subjects with low dS/dN ratios vs. subjects with high dS/dN ratios.
- Alignment between clones of subjects 4, 9, 11, 14 (dS/dN=0) was run. We observed these amino acid sequences to have: Strong:20 Weak:6 Conserved:47 Non-Consensus:22
- Alignment between clones of subjects 2, 5, 7, 13 (dS/dN>0) was run. We observed these amino acid sequences to have: Strong:22 Weak:10 Conserved:51 Non-Consensus:12
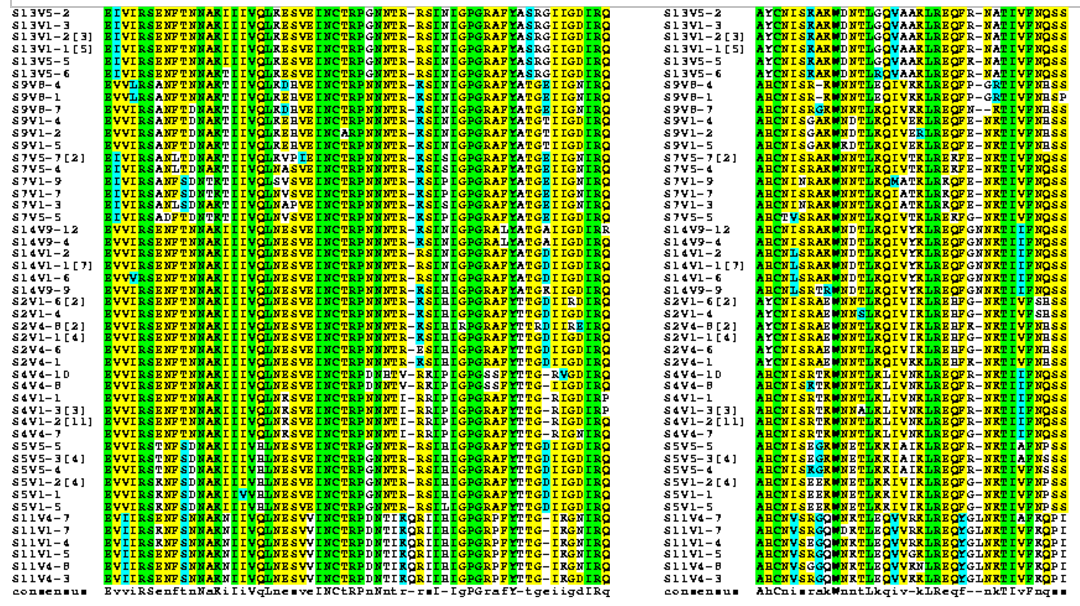
- Alignments were rerun using the BoxShade tool to help us see where changes in amino acids were occurring along the sequences.
- Using information from the Journal Club articles, we selected 6 amino acids along the V3 region and observed their specific changes.
- We used the Protein Predictor website to understand how severe these changes in amino acids are.
- Problem: protein they crystallized is not the same as in Markham
- Turning strong weak conserved and nonconsensus data into ratios may be better for comparison. 10 more positions where there are changes in rapid progressor group. We have count and position data...are there different patterns where the nonconsenesus spots occur along the gene? We can use the 3D model to show where similarities and difference are in these positions.
- Positions along the protein where the sequence alignment is highly variable across all subjects therefore defined as "non-consensus": 37, 41, 50, 79, 86, 87.
- We selected these positions on the 3D model to highlight where they are positioned along the protein.
Data and Files
HIV Structure Project
- Link to slides saved Monday 11/7/16: Media:Zach&JordanHIVRoughDraft.pdf
Conclusion
A rough draft of our powerpoint presentation was created this week. Many different programs were used to discover differences in the amino acids between different subjects. Amount of nonconsensual sites were found for both high dS/dN subjects and low dS/dN subjects. Six variable sites of interests were detected and analyzed using 3D imaging. A clearer picture of the variation in structure of HIV was formulated however it is still unclear how these changes relate to function
Acknowledgements
- I worked with my partner Zach Goldstein both in class and on Monday night on our powerpoint project. I also share both methods and data sections with him for this electronic notebook.
- I received instruction from Dr. Dahlquist's in class as well
- While I worked with the people noted above, this individual journal entry was completed by me and not copied from another source.
Jordan T. Detamore 23:27, 7 November 2016 (EST):
References
- Week 10 Assignment
- NCBI Open Reading Frame Finder
- ExPASY Translate tool
- PredictProtein server
- NCBI Structure Database
- Huang et al. (2005) structure 2B4C
- Cn3D software site
Useful links
LMU Seaver College of Science and Engineering