Isai Lopez Individual Journal 10
Purpose
To continue the work we started last week in lab and finish the exploration of the protein sequence analysis tools as well as performing further analysis to verify how amino acid changes affect the overall 3D structure of a molecule. In addition, we will be submitting our current work towards the powerpoint presentation for next week and displaying the latest data tables.
Methods/ Results
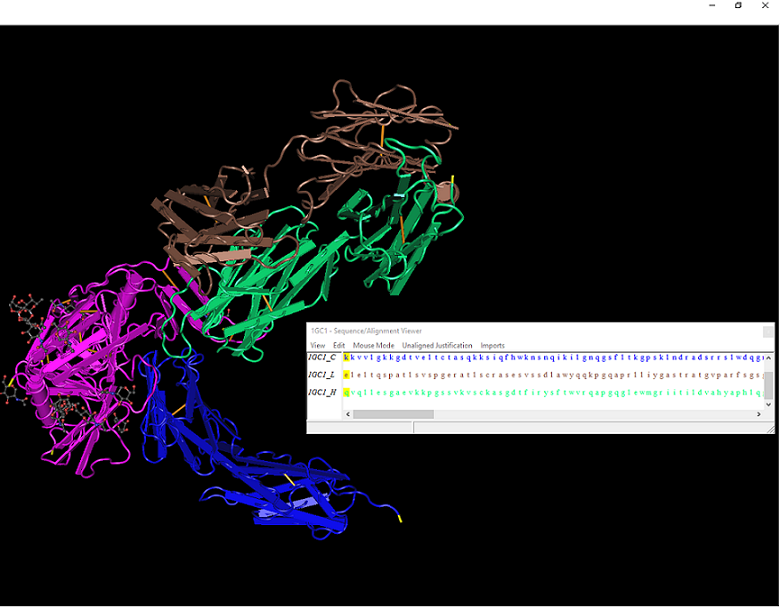
- The N and C termini are highlighted yellow in the picture. To find them, simply select the first and last amino acids from the sequence given by the Cn3D program.
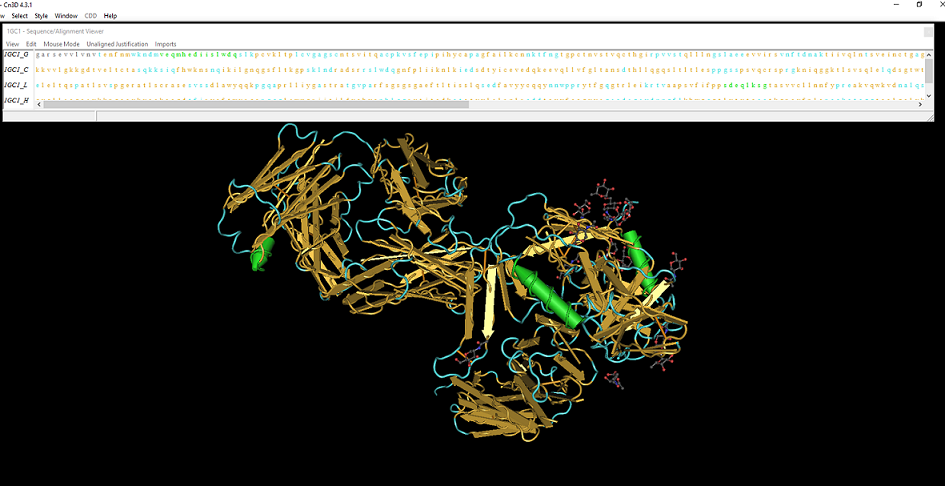
- To highlight the secondary structures for the complex given in the Kwong study, click the style tab, then coloring shortcuts, then select secondary structure. Highlighted in the picture below are the secondary structures for the 3D structure from Kwong et al. (1998). In yellow are the beta sheets, while in green are alpha helices within the complex. The predictions made by the PredictProtein server seem to match with those made by the Cn3D software for the gp120 molecule.
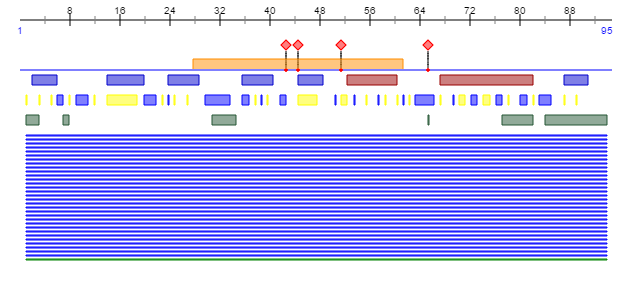
- The V3 structure is truncated and cut in the 3D version shown by the Kwong et al. paper, although the PredictProtein program shows the relative location of the V3 region bordered on both sides by disulfide bonds at the 28 and 62 residue positions in the Markham et al. paper.
- Now use the Biology Workbench and perform a sequence alignment for both the amino acid sequence and the DNA sequences of the data.
- More differences appear in the DNA sequences than in the amino acid sequences
- The reason for this is likely that the DNA sequence can code for the same amino acid in more than one combination, so differences in DNA will not necessarily effect amino acid sequence.
- Now analyze the amino acid sequences across the alignments you perform.
- The changes in amino acid would likely change the 3D structure of the protein in some way, given that some of the residues change to different families (basic>acidic, polar>nonpolar) which would likely change the interactions that that specific amino acid would have with other parts of the sequence, thus changing the overall protein structure.
Conclusion
During this week, I finished up the work we started in class last week. Using the Cn3D software, I was able to look at the amino acid sequence and pick out specific amino acid sequences that correspond to secondary structures as well as larger domains within the complex. Using the PredictProtein program, I was able to look at the "map" of the gp120 protein sequence and match it up with the 3D structure downloaded online to compare the two. I was also able to visualize how differences in amino acid family and type had an effect on the overall 3D structure using the software available. Using these tools, we can now proceed to analyze how differences in amino acid sequence for our chosen clones and subjects affect the 3D structure and use that to look at our project through a more focused lens.
Powerpoint Presentation
HIV Structure Research Project Progress Presentation
Data & Files
- http://openwetware.org/wiki/Image:IsaiLopezTerminalCN.png
- http://openwetware.org/wiki/Image:IsaiLopezSecondary.png
- http://openwetware.org/wiki/Image:HIVvariable3regionLopez.png
Acknowledgments
I worked outside of class with Colin Wikholm over Facebook messenger to prepare the powerpoint progress presentation. We also worked in class in terms of making sure each of us had finished each of the necessary components of the in-class activity. I also used the syntax for one of the links for the Kwong paper and 3D structure file download from the Week 10 Assignment Page written by Dr. Dahlquist