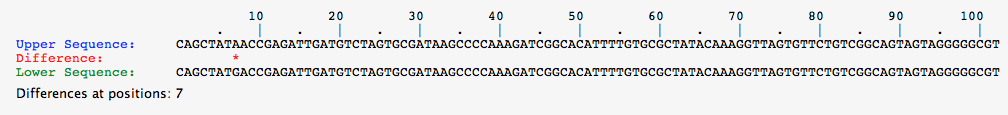Isai Lopez Individual Journal 2
Purpose
The purpose of this assignment was to use the Aipotu program to create a true breeding purple flower using models and principles of molecular biology.
Methods
- Use the steps outlined in the instruction packet for Part III: Molecular Biology
- To begin, open Aipotu and select the Molecular Biology tab in the top left of the window.
- Task I
- To complete this task, double-click the red organism in the greenhouse and select one of the nucleotides for the white allele (this should be in the lower gene window by default) and change it to a different base, then change it back.
- Click the "fold protein" button. This should add the allele to your history list.
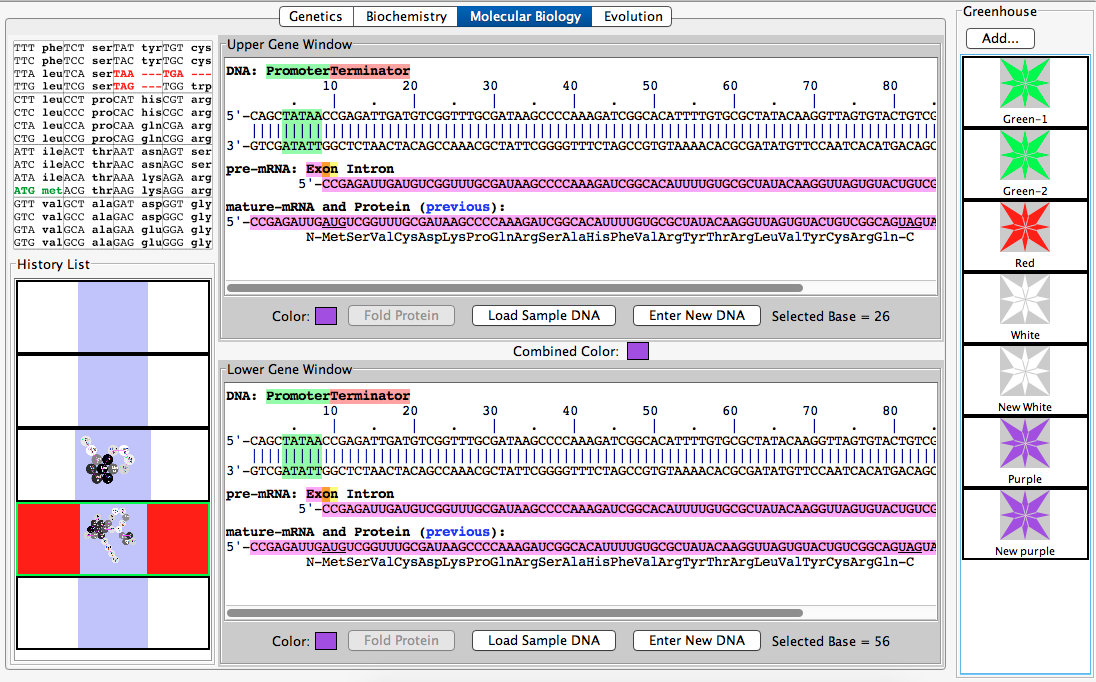
- Select the white organism from the greenhouse, which will populate both gene windows with white alleles (these two alleles are identical).
- Now double click your white allele from the history list and choose either "send to upper panel" or "send to lower panel", it will not matter.
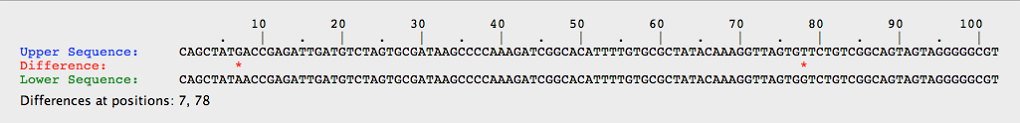
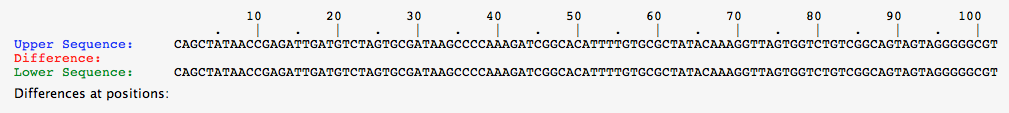
- Click the compare tab at the top of the page and compare the upper vs lower. This will tell you which nucleotides are different between the two white alleles.
- The figure above shows that two white alleles have differing DNA sequences.
- Task II
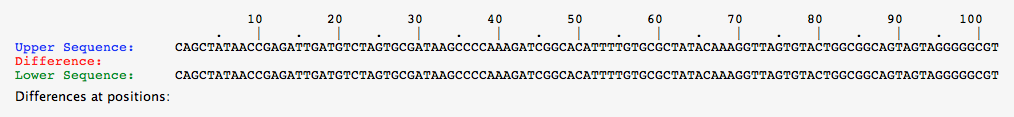
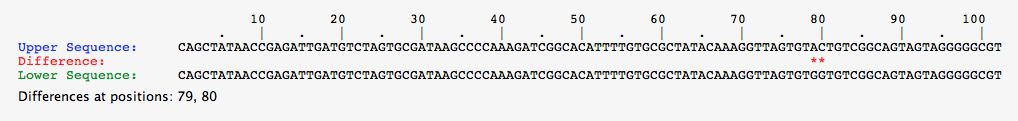
- For this task, select each organism from the greenhouse individually, and perform a comparison between the alleles shown.
- Below are the pictures for the comparisons for each organisms, note that if there is no red asterisk between the alleles, the DNA sequence is identical.
- Comparison for alleles of Green-1 organism
- Comparison for alleles of Green-2 organism
- Comparison for alleles of Red organism
- Comparison for alleles of White organism
- Task III
- For this task, select the Biochemistry tab and in the "Upper folding window", type the sequence for the protein "MLVKEIAMYRFATHER" exactly as shown. This should create a purple protein
- Now in the Molecular Biology tab, select "enter new DNA" type in any nucleotide sequence that corresponds to the amino acid sequence that codes for the protein above.
- Use the compare tab to show that the combination of the two alleles creates a purple flower, and that the combination will not create another color, thus showing that it is true-breeding.
Conclusion
Our main finding for today's project was a reverse engineered DNA sequence that yielded a true-breeding purple flower, starting with a protein. We fulfilled our purpose in that we were able to test that the plants coded for by the protein were both purple and true breeding when self crossed.
Acknowledgements
- During the course of this assignment, I worked with my group mates Matt Oki and Avery Vernon-Moore. We collaborated throughout class and communicated using email by sending pictures of our work.
- Isai Lopez 02:09, 13 September 2016 (EDT):
- I also received assistance from Dr. Dahlquist, Ph.D in class in the form of questions answered about the assignment.
- I used the syntax and link for my cited article from the bottom of the [Week 2] page originally written by Dr. Dahlquist, PhD.
- While I worked with the people noted above, this individual journal entry was completed by me and not copied from another source
References
For this assignment, I used the [Aipotu] website and program. The instructions for this assignment came from our [Week 2] web page I also used the journal article submitted by Dr. Dahlquist, Ph.D. The citation is as follows Eaton, C.D., Callendar, H.L., Dahlquist, K.D., LaMar, M.D., Ledder, G., Schugart, R.C. (2016) A “Rule of Five” Framework for Models and Modeling to Unify Mathematicians and Biologists and Improve Student Learning. Submitted on 20 June 2016 to the journal PRIMUS: Problems, Resources, and Issues in Mathematics Undergraduate Studies for an upcoming special issue on interdisciplinary conversations.