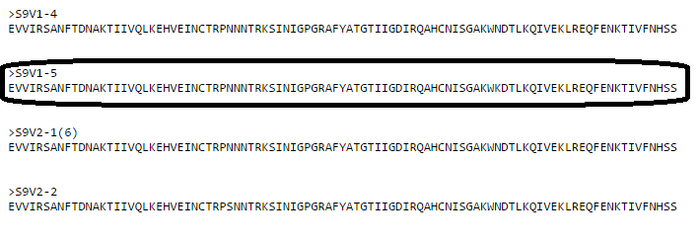ColinWikholm BIOL368 Week 9
Electronic Notebook Week 9
Purpose
The goal of this week's in-class work was to learn and utilize protein sequencing software to investigate the polypeptide-level structure of the gp120 glycoprotein of HIV-1 and prepare a relevant research project using these tools.
Methods and Results
- Use the ExPASy Translate Tool or NCBI Open Reading Frame Finder to convert a DNA sequence from the Markham et al. (1998) study into a protein sequence.
- Identify the correct reading frame (how do you know?)
- Six reading frames were outputted after inputting a nucleic acid sequence from HIV-1 clone S9V1-5 (see image below). The correct reading frame is 5'3' Frame 1 (outlined). Because the nucleic acid is a region within the gp120 gene, there should be no effects of stop codons in the correct reading frame. Because 5'3' Frame 1 is the only choice that doesn't show signs of a stop codon, it is thus the correct reading frame
- Confirm the above answer using the Bioequest BEDROCK Amino Acid Sequences
- The 5'3' Frame 1 output matched the BEDROCK sequence (image below), and is thus confirmed to be the correct reading frame:
- Identify the correct reading frame (how do you know?)
- Investigate current knowledge about the gp120 glycoprotein using the Universal Protein resource repository (UniProt)
- Use "HIV" and "gp120" as keywords to search the database. How many results are found?
- The search returned 206,278 results.
- Access the entry labeled with accession number "P04578" links to the gp120 repository information. What sort of information is provided about gp120 in this entry?
- Information included in the database for the gp120 protein included it's function in HIV-1, various names and its taxonomy, location in HIV-1, effects of some mutations in its nucleic acid sequence, protein modifications and components, interactions and structure, as well as detailed information about it's DNA sequence, useful related resources, and miscellaneous information.
- Use "HIV" and "gp120" as keywords to search the database. How many results are found?
- Access the free to use Open PredictProtein.
- Copy and paste a Markham et al. (1998) amino acid sequence as an entry and press submit.
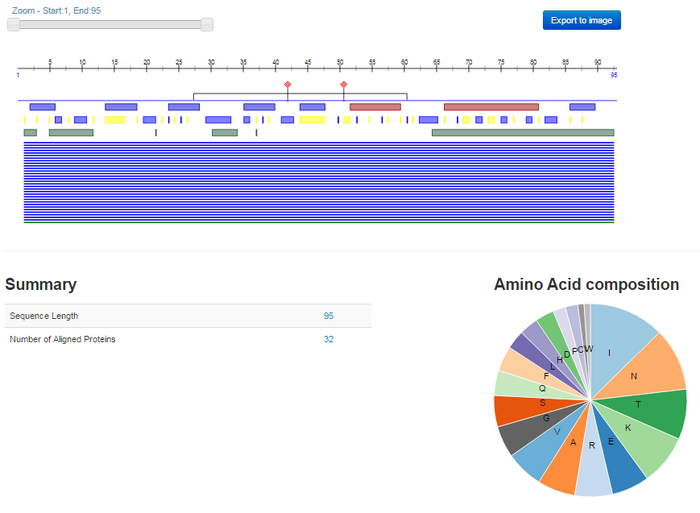
- The following visual output was given:
- What sort of information is provided? How does this relate to the information you found previously in UniProt?
- Like in UniProt, Open PredictProtein gives information on gp120 related to structure, function, and additional resources and related literature. However, Open PredictProtein gives information that is centered around diagramming, such as figures of amino acid sequences and charts of amino acid types. Although the information is not as extensive as that provided by UniPort, it is more concise and more useful for integrating information using visual conceptualization.
- Copy and paste a Markham et al. (1998) amino acid sequence as an entry and press submit.
Time ran out before the rest of the procedure could be completed in class. The methods can be found in the Week 9 Assignment.
Data and Files
Conclusion
Use of protein sequencing and analysis tools allowed for improved understanding and visualization of the the gp120 envelope protein. I discovered that the correct reading frame from the Markham et al. (1998) study would not show effects of a stop codon. I could use this information to identify the correct reading frame for further amino acid sequence analysis. Using various protein database and analysis softwares, I was able to visualize amino acid components of gp120 and learn more about its function, structure, and other important information. I was even able to visualize the effects of mutations on various parts of the gp120 (including the V3 region). These skills and tools gave me an improved understanding of how nucleic acid sequence affects amino acid sequence, and how this subsequently affects gp120 functioning and characteristics within HIV-1. After learning to do all of these things, I was able to formulate ideas and plans for my protein analysis project of gp120. The details of the project can be found below.
Defining You HIV Structure Research Project
- What is your question?
- How does degree of amino acid change due to mutations in the gp120 sequence from the Markham et al. (1998) study relate to rate of HIV-1 progression?
- Make a prediction about the answer to your question before you begin your analysis.
- We predict that a greater degree of amino acid change will be associated with a greater level of HIV-1 progression.
- Which subjects, visits, and clones will you use to answer your question? Justify why you chose the subjects, visits, and clones you did.
- We will use two clonal sequences from each of the 15 subjects at the beginning of the study and compare them to the same sequence in two clones for each of the 15 subjects at their last visit for the study. We chose to do this because it will ideally give a better overall view of how amino acid changes affect the seroconversion rate, while accounting for subjects in all three progression groups. This will give us a total of 60 sequences to work with, and while that is more than the ~50 recommended, we have used the alignment tool with around this many sequences in the past with success.
Acknowledgments
Isai Lopez and I worked in-person together during the Bioinformatics class period on October 24, 2016 to complete the in-class activity and make plans for our gp120 structure analysis project. We received assistance from Kam D. Dahlquist on reading frames during this time as well. Lastly, I gained wiki syntax information on indentations from Help:Wiki markup. While I worked with the people noted above, this individual journal entry was completed by me and not copied from another source.
Colin Wikholm 23:21, 31 October 2016 (EDT)
References
Important links
Bioinfomatics Lab: Fall 2016
Class Page: BIOL 368-01: Bioinfomatics Laboratory, Fall 2016
| Weekly Assignments | Individual Journal Assignments | Shared Journal Assignments |
|---|---|---|
|
|
|