ColinWikholm BIOL368 Week 6
HIV Evolution and Electronic Lab Notebook
Purpose
The env gene is important in the invasion, host evasion, and survival of HIV-1, so studying HIV-1 env sequences may offer the key to more effective HIV-1 treatments. The purpose of this week’s work was to perform the study described in Individual Journal 4 and expounded on in Individual Journal 5. We expected that re-analysis of the data from the Markham et al. (1998) study using different statistical methods would show a positive correlation between genetic divergence and rate of CD4 T cell decline.
Methods and Results
- Subjects 6, 8, 9, and 14 were selected for study for having the highest number of visit data points. This allowed for minimized uncertainty without using data from all 15 patients.
- Mia Huddleston, Anindita Varshneya, William Fuchs, and I each self-assigned one subject to use for calculations.
- Using Biology Workbench and ClustalW, the sequences were aligned. Phylogenetic trees were produced for the clones of each of the subjects: Rooted Phylogenetic Trees
- S was calculated for each of the patients for each of their visits by counting the number of base pair differences between each clone.
- Diversity (theta) of each subject was calculated using the procedure described in Week 4 Individual Journal - Activity 2, Part 2: Quantifying Diversity Within and Between Subjects.
- William Fuchs and I calculated divergence for each of the four subjects’ visits using the following formula: Divergence = ([|(θ2-θ1)/θ1|]/time).
- CD4 T cell data was collected of each of the four patients was collected from the Markham et al. 1998 data table.
- CD4 T cell data and divergence data were transferred to SPSS Statistics 21. All data analysis was performed on this program
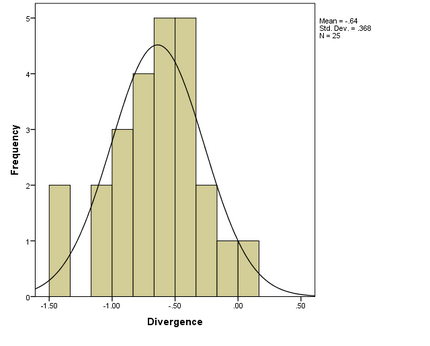
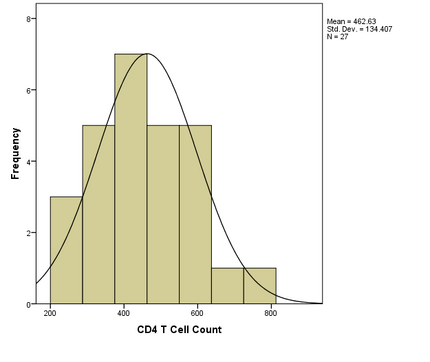
- CD4 T cell count and divergence data were tested for normality using visual analysis of histograms. The analysis showed normal distributions for both CD4 T cell count and divergence:
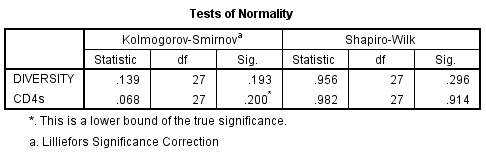
- Normality of CD4 T cell count and divergence was evaluated by the Shapiro-Wilk test for normality. This was used instead of the Kolmogorov-Smirnov test because n<50. The following was outputted by SPSS:
- A Pearson product-moment correlation coefficient was used to test for a correlation between CD4 T cell count and divergence. This was performed for each individual subject and for combined subject data.
- No test showed a significant correlation between divergence and CD4 T cell count, as evident in the Pearson test SPSS output.
- Scatter plots were created for each of the patients and for the combined data.
- The following scatter plots are supplements to the results of the Pearson test for normality: SPSS Scatterplot outputs. Note that the SPSS output does not include labels of the subjects, which is ordered in a descending fashion -6,8,9,14.
Data and Files
- HIV Evolution Project Presentation
- Markham et al. 1998 data table (taken from the Week 6 Assignment Page)
- Divergence and CD4 T cell count histograms
- Pearson test SPSS output
- SPSS Scatterplot outputs
- Rooted Phylogenetic Trees
Scientific Conclusion
We did not find that genetic divergence was positively correlated with CD4 T cell decline, and did not find significant correlations between the two variables in individual subjects or combined data. Our study was limited by time and resources. Consequently, we did not use the same measure of divergence used by Markham et al. (1998). Future studies should longer in time and would allow us to follow the same divergence described in Markham et al. (1998). Some other limitations of this study include limited sample size (we only analyzed subjects 6, 8, 9, and 14) and limited subject representation (subjects 6, 8, 9, and 14 were all from the moderate progressor group assigned in the Markham study). A final factor that may have influenced the results was that subject 9 was noted in the Markham study as having unusually high env diversity. Nonetheless, this would not have affected the correlation analysis of the individual subjects. For fairer reevaluation of the Markham study, the methods and sampling should be copied, with only the statistical tests changing.
Recent studies have confirmed, and also expanded on, the results of the Markham study. In the HIV-1 env gene, co-linear increase in genetic divergence and diversity, a plateau, and then sometimes a decrease in the two factors (Merk & Subramaniam 2013). The ability of an infectious agent to evade host immunity or live in harsh environments is not limited to HIV-1, and is seen in microbes of other serious disorders such as Cystic fibrosis (Oliver 2000). Studying the evolution of infectious agents is a promising avenue of research. We hope to investigating the data-set of Markham et al. to better understand HIV-1 and possibly other human pathogens.
HIV Evolution Powerpoint Presentation
The above scientific notebook work has been refined and assimilated into HIV Evolution Project Presentation. Many of the outputs above are the raw results from the software and are not the finalized results that were finalized within the presentation.
Acknowledgements
As stated in my Week 5 Individual Journal, I discussed project plans with Anindita Varshneya, Mia Huddleston, and William P Fuchs in class and in person on September 27, 2016. What is more, we all talked over Facebook Messenger on October 2, 2016 to reaffirm that we would be conducting diversity and divergence analysis separately (but assist each other if needed) during class on October 4, 2016. For the diversity component of the parallel projects, see Anindita Varshneya's procedure at AninditaVarshneya BIOL368 Week 5. I met with William P Fuchs in person to prepare for our project procedure on October 4, 2016. Since this time, I have been in contact most days with William P Fuchs over Facebook Messenger. We did not meet face-to-face while working on the Powerpoint Presentation during this time, but rather exchanged work over Messenger while I was out of state. For assitance with syntax, I used the Wiki Help List. Finally, Dr. Dahlquist reviewed my PowerPoint presentation over email, assisted in project setup and data analysis in class during weeks 4 and 5, as helped me make presentation arrangements out of class on October 4, 2016. While I worked with the people noted above, this individual journal entry was completed by me and not copied from another source.
Colin Wikholm 16:02, 10 October 2016 (EDT)
References
- CLUSTAL W: Julie D. Thompson, Desmond G. Higgins and Toby J. Gibson, modified; any errors are due to the modifications. PHYLIP: Felsenstein, J. 1993. PHYLIP (Phylogeny Inference Package) version 3.5c. Distributed by the author. Department of Genetics, University of Washington, Seattle.
- Evering, T. H., Kamau, E., St. Bernard, L., Farmer, C. B., Kong, X.-P., & Markowitz, M. (2014). Single genome analysis reveals genetic characteristics of Neuroadaptation across HIV-1 envelope. Retrovirology, 11, 65. http://doi.org/10.1186/s12977-014-0065-0
- Gall, A., Kaye, S., Hué, S., Bonsall, D., Rance, R., Baillie, G. J., … Kellam, P. (2013) Restriction of V3 region sequence divergence in the HIV-1 envelope gene during antiretroviral treatment in a cohort of recent seroconverters. Retrovirology, 10, 8. http://doi.org/10.1186/1742-4690-10-8\
- Higgins, D.G., Bleasby, A.J. and Fuchs, R. (1992) CLUSTAL V: improved software for multiple sequence alignment. Computer Applications in the Biosciences (CABIOS), 8(2):189-191. Thompson J.D., Higgins D.G., Gibson T.J. "CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice." Nucleic Acids Res. 22:4673-4680(1994). Felsenstein, J. 1989. PHYLIP -- Phylogeny Inference Package (Version 3.2). Cladistics 5: 164-166.
- Oliver, A., Cantón, R., Campo, P., Baquero, F., & Blázquez, J. (2000). High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science, 288(5469), 1251-1253.
- Markham, R. B., Wang, W.-C., Weisstein, A. E., Wang, Z., Munoz, A., Templeton, A.,… Yu, X.-F. (1998). Patterns of HIV-1 evolution in individuals with differing rates of CD4 T cell decline. Proceedings of the National Academy of Sciences of the United States of America, 95(21), 12568–12573.
- Mascola, J. R., & Montefiori, D. C. (2003). HIV-1: nature's master of disguise. Nature medicine, 9(4), 393-394.
- Merk, A., & Subramaniam, S. (2013). HIV-1 envelope glycoprotein structure. Current Opinion in Structural Biology, 23(2), 268–276. http://doi.org/10.1016/j.sbi.2013.03.007
- Müller, F. (2009). Assessing Antibody Neutralization of HIV-1 as an Initial Step in the Search for gp160-based Immunogens (Doctoral dissertation, Universität des Saarlandes Saarbrücken).
- Wang, W., Nie, J., Prochnow, C., Truong, C., Jia, Z., Wang, S., … Wang, Y. (2013). A systematic study of the N-glycosylation sites of HIV-1 envelope protein on infectivity and antibody-mediated neutralization. Retrovirology, 10, 14. http://doi.org/10.1186/1742-4690-10-14
Important links
Bioinfomatics Lab: Fall 2016
Class Page: BIOL 368-01: Bioinfomatics Laboratory, Fall 2016
| Weekly Assignments | Individual Journal Assignments | Shared Journal Assignments |
|---|---|---|
|
|
|