ColinWikholm BIOL368 Week 10
Electronic Notebook Week 10
Purpose
To learn and apply methods of protein structure visualization and analysis, and to continue work on the HIV-1 protein structure project.
Methods and Results
- Download the 1GC1 structure file found at Kwong et al. (1998) NCBI Webpage
- Use the Cn3D macromolecular structure viewer found at the Cn3D NCBI Webpage to open the file and perform the following:
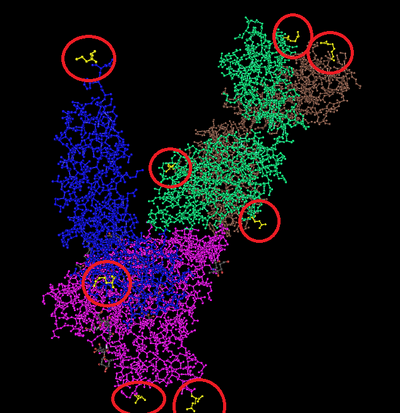
- Find the N-ter and C-ter for the tertiary structure of each polypeptide.
- The N-ter and C-ter are colored yellow and circled in red:
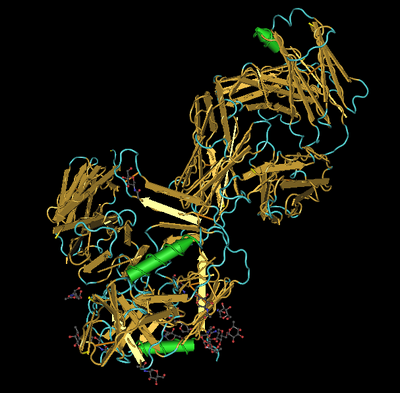
- Identify all secondary structures and report if they match the PredictProtein predictions
- The structure shows alpha-helices (green), beta-sheets (gold), and random coils (turquoise). They do match the PredictProtein predictions of secondary structure.
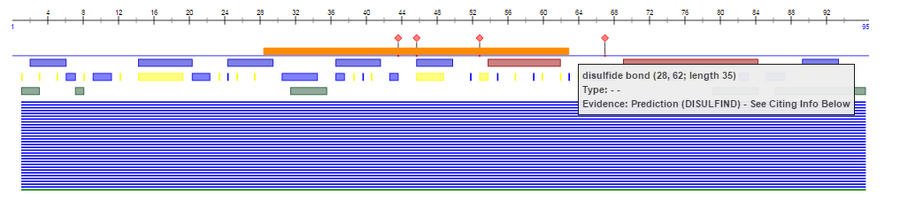
- Find the V3 region and identify the location that Markham et al. (1998) used for sequencing analysis.
- The V3 is truncated on the structure data from the Kwong et al. (1998), but the Predict Protein shows the V3 region (amino acids 296-330 of gp120) shown connected into a loop by a disulfide bond (below) on the gp120 sequence used in the Markham at al. .
- Find the N-ter and C-ter for the tertiary structure of each polypeptide.
- For the protein sequence your are analyzing, use Biology Workbench to perform sequence alignment
- Are there more or less differences between the aligned DNA sequences or amino acid sequences?
- There are more differences between the DNA sequences than the amino acid sequences.
- Why is this?
- The genetic code is degenerate, as more than one codon often codes for the same amino acid. There may be differences in DNA sequence without changes in the amino acid sequence.
- Are there more or less differences between the aligned DNA sequences or amino acid sequences?
- Analyze the multiple sequence alignment. Do you think the observed amino acid changes would affect the 3D structure? Explain.
- Many of the amino acid changes are from one amino acid classification to another, so I believe that the changes would affect the 3D structure. This is further supported by the Markham et al. (1998) study, which showed these differences as affecting evasion of HIV-1 from the host immune system. If there is a change in functionality, then there is likely a change in structure.
- Prepare an 5-6 minute informal presentation of your project progress using a format similar to Markham et al. (1998) presentation directions
- The powerpoint of the project progress can be found in HIV Structure Research Project Progress
Data and Files
- HIV Structure Research Project Progress Presentation
- N-ter and C-ter Image
- Secondary Structures Image
- PredictProtein Markham et al. (1998) Sequence Image
Conclusion
I successfully learned to use protein sequencing and analysis tools, and also applied them to advance our investigation of the structure of HIV-1 gp120. Applying each of these skills, the protein sequences were aligned and analyzed for strong, weak, no consensus, and fully conserved regions. I now have the ability to analyze and visualize protein sequences, as well as compare them using multiple sequence alignment. What is more, analyzing HIV-1 components and interactions has allowed me to interpret structure and its relationship to function. This will be highly applicable for continuation of the the HIV-1 protein structure project, and also offers further areas of interest, such as differing methods of consensus or mutation analysis with respect to amino acid evolutionary similarity.
HIV Structure Research Project Progress
HIV Structure Research Project Progress Presentation
Acknowledgments
I worked with Isai Lopez in class to perform the in-class assignment, as well as to sequence and analyze the protein data for the project. We also performed data analysis and prepared the informal presentation together on a daily basis in person outside of class. During November 5-7, 2016, we worked on the presentation together over Facebook messenger. I would also like to thank Kam D. Dahlquist for her in-class assistance with navigating the software during week 10. While I worked with the people noted above, this individual journal entry was completed by me and not copied from another source.
Colin Wikholm 01:56, 8 November 2016 (EST)
References
Important links
Bioinfomatics Lab: Fall 2016
Class Page: BIOL 368-01: Bioinfomatics Laboratory, Fall 2016
| Weekly Assignments | Individual Journal Assignments | Shared Journal Assignments |
|---|---|---|
|
|
|