Aiden Burnett Week 4
From OpenWetWare
Jump to navigationJump to search
Purpose
Gain experience obtaining sequence data, comparing it using multiple sequence alignments, and analyzing it for phylogenetic relationships using trees. This should equip me to answer any questions I may have by using these tools.
Methods & Results
GenBank
- I Chose one of the GenBank records from the Data & Resources section of the week 3 assignment page and viewed both the full record and the FASTA formatted sequence.
- I chose "Severe acute respiratory syndrome coronavirus 2 isolate Wuhan-Hu-1"
- accession number: MN908947
- The GenBank record provides the virus's genetic sequence (bases 1 to 29903), as well as information identifying organism & the source paper.
- I downloaded the nucleotide sequence in FASTA format to my local hard drive.
- This was done by clicking the "Send to" link in the upper right of the page. I then selected "Complete Record", File as the Destination, and FASTA as the format. I clicked the "Create File" button. (Being careful to remember where you put the file and what you name it so that you can find it later.)
- I opened the file with a word processor to confirm that I had the sequence and that it is in the FASTA format.#
- I chose "Severe acute respiratory syndrome coronavirus 2 isolate Wuhan-Hu-1"
- I was assigned the nucleotide sequence accession number KC881006 from Figure 2 of the Wan et al. paper.
- I searched for the GenBank record associated with this sequence, as well as the spike protein specifically. I then added a hyperlink to the GenBank record for these sequences in the Data & Tools section of assignment 4.
- I then added the protein sequence below to the talk page for this assignment.
>AGZ48818.1 spike protein [Bat SARS-like coronavirus Rs3367] MKLLVLVFATLVSSYTIEKCLDFDDRTPPANTQFLSSHRGVYYPDDIFRSNVLHLVQDHFLPFDSNVTRF ITFGLNFDNPIIPFKDGIYFAATEKSNVIRGWVFGSTMNNKSQSVIIMNNSTNLVIRACNFELCDNPFFV VLKSNNTQIPSYIFNNAFNCTFEYVSKDFNLDLGEKPGNFKDLREFVFRNKDGFLHVYSGYQPISAASGL PTGFNALKPIFKLPLGINITNFRTLLTAFPPRPDYWGTSAAAYFVGYLKPTTFMLKYDENGTITDAVDCS QNPLAELKCSVKSFEIDKGIYQTSNFRVAPSKEVVRFPNITNLCPFGEVFNATTFPSVYAWERKRISNCV ADYSVLYNSTSFSTFKCYGVSATKLNDLCFSNVYADSFVVKGDDVRQIAPGQTGVIADYNYKLPDDFTGC VLAWNTRNIDATQTGNYNYKYRSLRHGKLRPFERDISNVPFSPDGKPCTPPAFNCYWPLNDYGFYITNGI GYQPYRVVVLSFELLNAPATVCGPKLSTDLIKNQCVNFNFNGLTGTGVLTPSSKRFQPFQQFGRDVSDFT DSVRDPKTSEILDISPCSFGGVSVITPGTNTSSEVAVLYQDVNCTDVPVAIHADQLTPSWRVYSTGNNVF QTQAGCLIGAEHVDTSYECDIPIGAGICASYHTVSSLRSTSQKSIVAYTMSLGADSSIAYSNNTIAIPTN FSISITTEVMPVSMAKTSVDCNMYICGDSTECANLLLQYGSFCTQLNRALSGIAVEQDRNTREVFAQVKQ MYKTPTLKDFGGFNFSQILPDPLKPTKRSFIEDLLFNKVTLADAGFMKQYGECLGDINARDLICAQKFNG LTVLPPLLTDDMIAAYTAALVSGTATAGWTFGAGAALQIPFAMQMAYRFNGIGVTQNVLYENQKQIANQF NKAISQIQESLTTTSTALGKLQDVVNQNAQALNTLVKQLSSNFGAISSVLNDILSRLDKVEAEVQIDRLI TGRLQSLQTYVTQQLIRAAEIRASANLAATKMSECVLGQSKRVDFCGKGYHLMSFPQAAPHGVVFLHVTY VPSQERNFTTAPAICHEGKAYFPREGVFVFNGTSWFITQRNFFSPQIITTDNTFVSGSCDVVIGIINNTV YDPLQPELDSFKEELDKYFKNHTSPDVDLGDISGINASVVNIQKEINRLNEVAKNLNESLIDLQELGKYE QYIKWPWYVWLGFIAGLIAIVMVTILLCCMTSCCSCLKGACSCGSCCKFDEDDSEPVLKGVKLHYT
Phylogeny
- In my browser, I went to the website [www.phylogeny.fr.] & scrolled down on the page to the section labeled ‘Phylogeny analysis’, and then clicked on the text ‘One Click’.
- I clicked in the large text field labeled ‘Upload your set of sequences in FASTA, EMBL, or NEXUS format’ & copied the list of sequences from the talk page, using Ctrl-V to paste my sequences here, then clicking the “Submit” button.
- I pasted my sequences into the notepad application to ensure that it was simple text being fed into the phylogeny program.
- I navigated to the page named Alignment results. After the alignment was complete, a new page named Phylogeny phylogeny appeared. Lastly, a page named Tree rendering results appeared. I came back to these pages later. At this point I found the numbered tabs located just beneath the text One Click Mode, and click on the tab labeled 3. Alignment.
- Within the alignment, individual positions were color-coded to indicate their conservation, or how similar the sequences are to each other at that position. Blue highlighting indicated high conservation, while gray highlighting indicated lower conservation and white highlighting indicated little if any conservation.
- Near the bottom of the page, under Outputs, click on Alignment in Clustal format. This displayed my alignment in a text-only format in which each position's conservation was indicated by a symbol underneath the alignment block (“*” for invariant, “:” for highly conserved, “.” for weakly conserved, and a space for not conserved). I copy and pasted this entire alignment into my individual journal entry below.
QDF43825.1 ---------MKLLVLV-----FATLVSSYTIEKCTDFD------DRTPPSNTQFLSSHRG
AGZ48818.1 ---------MKLLVLV-----FATLVSSYTIEKCLDFD------DRTPPANTQFLSSHRG
ALK02457.1 ----------MFIFLF-----FLTLTSGSDLESCTTFD------DVQAPNYPQHSSSRRG
AAS10463.1 ----------MFIFLL-----FLTLTSGSDLDRCTTFD------DVQAPNYTQHTSSMRG
AAP13441.1 ----------MFIFLL-----FLTLTSGSDLDRCTTFD------DVQAPNYTQHTSSMRG
AAP13567.1 ----------MFIFLL-----FLTLTSGSDLDRCTTFD------DVQAPNYTQHTSSMRG
QHD43416.1 ----------MFVFLV-----LLPLVSSQ----CVNLT------TRTQLPPAYTNSFTRG
AVP78031.1 -----------MLFFL-----FLQFALVN--SQCVNLT------GRTPLNPNYTNSSQRG
ABD75323.1 --------MKILIFAF-----LVTLVKAQ--EGCGVIN------LRTQPKLTQVSSSRRG
QDF43835.1 --------MKVLIVLL-----CLGLVTAQ--DGCGHIS------TKPQPLLDKFSSSRRG
ABD75332.1 --------MKVLIFAL-----LFSLAKAQ--EGCGIIS------RKPQPKMEKVSSSRRG
QDF43820.1 --------MKILIFAF-----LVTLVEAQ--EGCGIIS------RKPQPKMAQVSSSRRG
AAZ67052.1 --------MKILILAF-----LASLAKAQ--EGCGIIS------RKPQPKMAQVSSSRRG
AFS88936.1 ----MIHSVFLLMFLLTPTESYVDVGPDSVKSACIEVDIQQTFFDKTWPRPIDVSKA-DG
YP_0010399 MTLLMCLLMSLLIFVRGCDSQFVDMSPASNTSECLESQVDAAAFSKLMWPYPIDPSKVDG
::. . * . *
QDF43825.1 VYYPDDIFRSNVLHLVQDHFLPFDSNVTRFITFGLN-------------FDN---PIIPF
AGZ48818.1 VYYPDDIFRSNVLHLVQDHFLPFDSNVTRFITFGLN-------------FDN---PIIPF
ALK02457.1 VYYPDEIFRSDTLYLTQDLFLPFYSNVTGFHTINHR-------------FDN---PVIPF
AAS10463.1 VYYPDEIFRSDTLYLTQDLFLPFYSNVTGFHTINHT-------------FDD---PVIPF
AAP13441.1 VYYPDEIFRSDTLYLTQDLFLPFYSNVTGFHTINHT-------------FGN---PVIPF
AAP13567.1 VYYPDEIFRSDTLYLTQDLFLPFYSNVTGFHTINHT-------------FDN---PVIPF
QHD43416.1 VYYPDKVFRSSVLHSTQDLFLPFFSNVTWFHAIHVS------GTNGTKRFDN---PVLPF
AVP78031.1 VYYPDTIYRSDTLVLSQGYFLPFYSNVSWYYSLTTN-------NAATKRTDN---PILDF
ABD75323.1 VYYNDDIFRSDVLHLTQDYFLPFHSNLTQYFSLNIE-------SDKIVYFDN---PILKF
QDF43835.1 VYYNDDIFRSDVLHLTQDYFLPFDTNLTRYLSFNMD-------SATKVYFDN---PTLPF
ABD75332.1 VYYNDDIFRSDVLHLTQDYFLPFDSNLTQYFSLNID-------SNKYTYFDN---PILDF
QDF43820.1 VYYNDDIFRSDVLHLTQDYFLPFDSNLTQYFSLNVD-------SDRYTYFDN---PILDF
AAZ67052.1 VYYNDDIFRSNVLHLTQDYFLPFDSNLTQYFSLNVD-------SDRFTYFDN---PILDF
AFS88936.1 IIYPQGRTYSNITITYQGLF-PYQGDHGDMYVYSAG--HATGTTPQKLFVANYSQDVKQF
YP_0010399 IIYPLGRTYSNITLAYTGLF-PLQGDLGSQYLYSVSHAVGHDGDPTKAYISNYSLLVNDF
: * *. . * * : : *
QDF43825.1 RDGVYF----AATEKSNVIRG-------------WVFGSTMNNKSQ---------SVIIM
AGZ48818.1 KDGIYF----AATEKSNVIRG-------------WVFGSTMNNKSQ---------SVIIM
ALK02457.1 KDGVYF----AATEKSNVVRG-------------WVFGSTMNNKSQ---------SVIII
AAS10463.1 KDGIYF----AATEKSNVVRG-------------WVFGSTMNNKSQ---------SVIII
AAP13441.1 KDGIYF----AATEKSNVVRG-------------WVFGSTMNNKSQ---------SVIII
AAP13567.1 KDGIYF----AATEKSNVVRG-------------WVFGSTMNNKSQ---------SVIII
QHD43416.1 NDGVYF----ASTEKSNIIRG-------------WIFGTTLDSKTQ---------SLLIV
AVP78031.1 KDGIYF----AATEHSNIIRG-------------WIFGTTLDNTSQ---------SLLIV
ABD75323.1 GDGVYF----AATEKSNVIRG-------------WVFGSTFDNTTQ---------SAIIV
QDF43835.1 GDGIYF----AATEKSNVVRG-------------WIFGSTMDNTTQ---------SAIIV
ABD75332.1 GDGVYF----AATEKSNVIRG-------------WIFGSSFDNTTQ---------SAIIV
QDF43820.1 GDGVYF----AATEKSNVIRG-------------WIFGSTFDNTTQ---------SAVIV
AAZ67052.1 GDGVYF----AATEKSNVIRG-------------WIFGSTFDNTTQ---------SAVIV
AFS88936.1 ANGFVVRIGAAANSTGTVIISPSTSATIRKIYPAFMLGSSVGNFSDGKMGRFFNHTLVLL
YP_0010399 DNGFVVRIGAAANSTGTIVISPSVNTKIKKAYPAFILGSSLTNTSAGQ-PLYANYSLTII
:*. . *:.. ..:: . :::*::. . : : ::
QDF43825.1 NNSTNLVIRACNFELCDNPFFVVLRSNNTQIPSY------IFNNAFN-CTFEYVSKDFNL
AGZ48818.1 NNSTNLVIRACNFELCDNPFFVVLKSNNTQIPSY------IFNNAFN-CTFEYVSKDFNL
ALK02457.1 NNSTNVVIRACNFELCDNPFFAVSKPTGTQTHTM------IFDNAFN-CTFEYISDSFSL
AAS10463.1 NNSTNVVIRACNFELCDNPFFVVSKPMGTRTHTM------IFDNAFN-CTFEYISDAFSL
AAP13441.1 NNSTNVVIRACNFELCDNPFFAVSKPMGTQTHTM------IFDNAFN-CTFEYISDAFSL
AAP13567.1 NNSTNVVIRACNFELCDNPFFAVSKPMGTQTHTM------IFDNAFN-CTFEYISDAFSL
QHD43416.1 NNATNVVIKVCEFQFCNDPFLGVYY--HKNNKSWMESEFRVYSSANN-CTFEYVSQPFLM
AVP78031.1 NNATNVIIKVCNFDFCYDP-YLSGY--YHNNKTWSIREFAVYSSYAN-CTFEYVSKSFML
ABD75323.1 NNSTHIIIRVCYFNLCKDPMYTVSA--GTQKSSW------VYQSAFN-CTYDRVEKSFQL
QDF43835.1 NNSTHIIIRVCYFNLCKEPMYAISN--EQHYKSW------VYQNAYN-CTYDRVEQSFQL
ABD75332.1 NNSTHIIIRVCNFNLCKEPMYTVSK--GTQQSSW------VYQSAFN-CTYDRVEKSFQL
QDF43820.1 NNSTHIIIRVCNFNLCKEPMYTVSR--GTQQSSW------VYQSAFN-CTYDRVERSFQL
AAZ67052.1 NNSTHIIIRVCNFNLCKEPMYTVSR--GAQQSSW------VYQSAFN-CTYDRVEKSFQL
AFS88936.1 PDGCGTLLRAFYCIL--EPRSGNHCPAGNSYTSF-----ATYHTPATDCSDGNYNRNASL
YP_0010399 PDGCGTVLHAFYCIL--KPRTVNRCPSGTGYVSY-----FIYETVHNDCQ-STINRNASL
:. ::.. : .* : : . . * . :
QDF43825.1 DIGEKPGNFKDLREFVFRNKDG--------FLHVYSGYQPISAASGLPTGF--NALKPIF
AGZ48818.1 DLGEKPGNFKDLREFVFRNKDG--------FLHVYSGYQPISAASGLPTGF--NALKPIF
ALK02457.1 DVAEKSGNFKHLREFVFKNKDG--------FLYVYKGYQPIDVVRDLPSGF--NILKPIF
AAS10463.1 DVSEKSGNFKHLREFVFKNKDG--------FLYVYKGYQPIDVVRDLPSGF--NTLKPIF
AAP13441.1 DVSEKSGNFKHLREFVFKNKDG--------FLYVYKGYQPIDVVRDLPSGF--NTLKPIF
AAP13567.1 DVSEKSGNFKHLREFVFKNKDG--------FLYVYKGYQPIDVVRDLPSGF--NTLKPIF
QHD43416.1 DLEGKQGNFKNLREFVFKNIDG--------YFKIYSKHTPINLVRDLPQGF--SALEPLV
AVP78031.1 NISGNGGLFNTLREFVFRNVDG--------HFKIYSKFTPVNLNRGLPTGL--SVLQPLV
ABD75323.1 DTSPKTGNFTDLREFVFKNRDG--------FFTAYQTYTPVNLLRGLPSGL--SVLKPIL
QDF43835.1 DTAPQTGNFKDLREYVFKNKDG--------FLSVYNAYSPIDIPRGLPVGF--SVLKPIL
ABD75332.1 DTAPKTGNFKDLREYVFKNKGG--------FLRVYQTYTAVNLPRGFPAGF--SVLRPIL
QDF43820.1 DTAPKTGNFKDLREYVFKNRDG--------FLSVYQTYTAVNLPRGLPIGF--SVLRPIL
AAZ67052.1 DTAPKTGNFKDLREYVFKNRDG--------FLSVYQTYTAVNLPRGLPIGF--SVLRPIL
AFS88936.1 NSFKE---YFNLRNCTFMYTYNITEDEILEWFGITQTAQGVHLFSSRYVDLYGGNMFQFA
YP_0010399 NSFK---SFFDLVNCTFFNSWDITADETKEWFGITQDTQGVHLYSSRKGDLYGGNMFRFA
: : * : .* . : . : . .: . : :
QDF43825.1 KLPLGINITNFRTLLTAF------PPNPGYWGTSAAAYFVGYLKPTTFMLKYDENGTITD
AGZ48818.1 KLPLGINITNFRTLLTAF------PPRPDYWGTSAAAYFVGYLKPTTFMLKYDENGTITD
ALK02457.1 KLPLGINITNFRAILTAF------LPAQDTWGTSAAAYFVGYLKPATFMLKYDENGTITD
AAS10463.1 KLPLGINITNFRAILTAF------SPAQDTWGTSAAAYFVGYLKPTTFMLKYDENGTITD
AAP13441.1 KLPLGINITNFRAILTAF------SPAQDIWGTSAAAYFVGYLKPTTFMLKYDENGTITD
AAP13567.1 KLPLGINITNFRAILTAF------SPAQDTWGTSAAAYFVGYLKPTTFMLKYDENGTITD
QHD43416.1 DLPIGINITRFQTLLALHRSYLTPGDSSSGWTAGAAAYYVGYLQPRTFLLKYNENGTITD
AVP78031.1 ELPVSINITKFRTLLTIHRGD---PMPNNGWTAFSAAYFVGYLKPRTFMLKYNENGTITD
ABD75323.1 KLPFGINITSFRVVMAMF------SKTTSNYVPESAAYYVGNLKQSTFMLSFNQNGTIVD
QDF43835.1 KLPIGINITSFKVVMSMF------SRTTSNFLPEVAAYFVGNLKYSTFMLNFNENGTITD
ABD75332.1 KLPFGINITSYRVVMTMF------SQFNSNFLPESAAYYVGNLKYTTFMLSFNENGTITD
QDF43820.1 KLPFGINITSYRVVMAMF------SQTTSNFLPESAAYYVGNLKYTTFMLRFNENGTITD
AAZ67052.1 KLPFGINITSYRVVMAMF------SQTTSNFLPESAAYYVGNLKYTTFMLSFNENGTITN
AFS88936.1 TLPVYDTIKYYSIIPHSIRSI---QSDRKAW----AAFYVYKLQPLTFLLDFSVDGYIRR
YP_0010399 TLPVYEGIKYYTVIPRSFRSK---ANKREAW----AAFYVYKLHQLTYLLDFSVDGYIRR
**. *. : : : **::* *: *::* :. :* *
QDF43825.1 AVDCSQNPLAELKCSVKSFEIDKGIYQTSNFRVAPSKEVVRFPNITNLCPFGEVFNATTF
AGZ48818.1 AVDCSQNPLAELKCSVKSFEIDKGIYQTSNFRVAPSKEVVRFPNITNLCPFGEVFNATTF
ALK02457.1 AVDCSQNPLAELKCSVKSFEIDKGIYQTSNFRVAPSKEVVRFPNITNLCPFGEVFNATTF
AAS10463.1 AVDCSQNPLAELKCSVKSFEIDKGIYQTSNFRVVPSGDVVRFPNITNLCPFGEVFNATKF
AAP13441.1 AVDCSQNPLAELKCSVKSFEIDKGIYQTSNFRVVPSGDVVRFPNITNLCPFGEVFNATKF
AAP13567.1 AVDCSQNPLAELKCSVKSFEIDKGIYQTSNFRVVPSGDVVRFPNITNLCPFGEVFNATKF
QHD43416.1 AVDCALDPLSETKCTLKSFTVEKGIYQTSNFRVQPTESIVRFPNITNLCPFGEVFNATRF
AVP78031.1 AVDCALDPLSETKCTLKSLTVQKGIYQTSNFRVQPTQSVVRFPNITNVCPFHKVFNATRF
ABD75323.1 AVDCSQDPLAELKCTTKSFNVSKGIYQTSNFRVSPVTEVVRFPNITNLCPFDKVFNATRF
QDF43835.1 AIDCAQNPLSELKCTIKNFNVSKGIYQTSNFRVSPTHEVIRFPNITNRCPFDKVFNASRF
ABD75332.1 AVDCSQNPLAELKCTIKNFNVSKGIYQTSNFRVTPTQEVVRFPNITNRCPFDKVFNASRF
QDF43820.1 AIDCAQNPLAELKCTIKNFNVSKGIYQTSNFRVSPTQEVVRFPNITNRCPFDKVFNASRF
AAZ67052.1 AIDCAQNPLAELKCTIKNFNVSKGIYQTSNFRVSPTQEVIRFPNITNRCPFDKVFNATRF
AFS88936.1 AIDCGFNDLSQLHCSYESFDVESGVYSVSSFEAKPSGSVVEQAEGVE-CDFSPLLSGTP-
YP_0010399 AIDCGHDDLSQLHCSYTSFEVDTGVYSVSSYEASATGTFIEQPNATE-CDFSPMLTGVA-
*:**. : *:: :*: .: :..*:*..*.: . . .: .: .: * * ::..
QDF43825.1 PSVYAWERKRISNCVADYSVLYNSTSFSTFKCYGVSATKLNDLCFSNVYADSFVVKGDDV
AGZ48818.1 PSVYAWERKRISNCVADYSVLYNSTSFSTFKCYGVSATKLNDLCFSNVYADSFVVKGDDV
ALK02457.1 PSVYAWERKRISNCVADYSVLYNSTSFSTFKCYGVSATKLNDLCFSNVYADSFVVKGDDV
AAS10463.1 PSVYAWERKRISNCVADYSVLYNSTSFSTFKCYGVSATKLNDLCFSNVYADSFVVKGDDV
AAP13441.1 PSVYAWERKKISNCVADYSVLYNSTFFSTFKCYGVSATKLNDLCFSNVYADSFVVKGDDV
AAP13567.1 PSVYAWERKKISNCVADYSVLYNSTFFSTFKCYGVSATKLNDLCFSNVYADSFVVKGDDV
QHD43416.1 ASVYAWNRKRISNCVADYSVLYNSASFSTFKCYGVSPTKLNDLCFTNVYADSFVIRGDEV
AVP78031.1 PSVYAWERTKISDCIADYTVFYNSTSFSTFKCYGVSPSKLIDLCFTSVYADTFLIRFSEV
ABD75323.1 PSVYAWERTKISDCVADYTVFYNSTSFSTFNCYGVSPSKLIDLCFTSVYADTFLIRFSEV
QDF43835.1 PNVYAWERTKISDCVADYTVLYNSTSFSTFKCYGVSPSKLIDLCFTSVYADTFLIRSSEV
ABD75332.1 PNVYAWERTKISDCVADYTVLYNSTSFSTFKCYGVSPSKLIDLCFTSVYADTFLIRSSEV
QDF43820.1 PNVYAWERTKISDCVADYTVLYNSTSFSTFKCYGVSPSKLIDLCFTSVYADTFLIRSSEV
AAZ67052.1 PNVYAWERTKISDCVADYTVLYNSTSFSTFKCYGVSPSKLIDLCFTSVYADTFLIRSSEV
AFS88936.1 PQVYNFKRLVFTNCNYNLTKLLSLFSVNDFTCSQISPAAIASNCYSSLILDYFSYPLSMK
YP_0010399 PQVYNFKRLVFSNCNYNLTKLLSLFAVDEFSCNGISPDSIARGCYSTLTVDYFAYPLSMK
..** ::* :::* : : : . .. *.* :*. : *::.: * * .
QDF43825.1 RQIAPGQTGVIADYNYKLPDDFMGC-VLAWNTRNIDATSTGNYNYKYRSLRHGKLRPFER
AGZ48818.1 RQIAPGQTGVIADYNYKLPDDFTGC-VLAWNTRNIDATQTGNYNYKYRSLRHGKLRPFER
ALK02457.1 RQIAPGQTGVIADYNYKLPDDFTGC-VLAWNTRNIDATQTGNYNYKYRSLRHGKLRPFER
AAS10463.1 RQIAPGQTGVIADYNYKLPDDFMGC-VLAWNTRNIDATSTGNYNYKYRYLRHGKLRPFER
AAP13441.1 RQIAPGQTGVIADYNYKLPDDFMGC-VLAWNTRNIDATSTGNYNYKYRYLRHGKLRPFER
AAP13567.1 RQIAPGQTGVIADYNYKLPDDFMGC-VLAWNTRNIDATSTGNYNYKYRYLRHGKLRPFER
QHD43416.1 RQIAPGQTGKIADYNYKLPDDFTGC-VIAWNSNNLDSKVGGNYNYLYRLFRKSNLKPFER
AVP78031.1 RQVAPGQTGVIADYNYKLPDDFTGC-VIAWNTAKQD---VGNYF--YRSHRSTKLKPFER
ABD75323.1 RQVAPGQTGVIADYNYKLPDDFTGC-VIAWNTAKQD---VGSYF--YRSHRSSKLKPFER
QDF43835.1 RQVAPGETGVIADYNYKLPDDFTGC-VIAWNTAKQD---QGQYY--YRSSRKTKLKPFER
ABD75332.1 RQVAPGETGVIADYNYKLPDDFTGC-VIAWNTAQQD---QGQYY--YRSYRKEKLKPFER
QDF43820.1 RQVAPGETGVIADYNYKLPDDFTGC-VIAWNTAKQD---TGHYY--YRSHRKTKLKPFER
AAZ67052.1 RQVAPGETGVIADYNYKLPDDFTGC-VIAWNTAKQD---QGQYY--YRSHRKTKLKPFER
AFS88936.1 SDLSVSSAGPISQFNYKQSFSNPTC-LILATVPHNLTTITKPLKYSYINKCSRLLSDDRT
YP_0010399 SYIRPGSAGNIPLYNYKQSFANPTCRVMASVLANVTITKPHAYG--YIS-KCSRLTGANQ
: ..:* *. :*** . * :: : * *
QDF43825.1 DISNVPFSPDGKPCTPP-AF-NCYW-----------PLNDYGFFTTNGIGYQPYRVVVLS
AGZ48818.1 DISNVPFSPDGKPCTPP-AF-NCYW-----------PLNDYGFYITNGIGYQPYRVVVLS
ALK02457.1 DISNVPFSPDGKPCTPP-AF-NCYW-----------PLNDYGFYITNGIGYQPYRVVVLS
AAS10463.1 DISNVPFSPDGKPCTPP-AP-NCYW-----------PLNGYGFYTTSGIGYQPYRVVVLS
AAP13441.1 DISNVPFSPDGKPCTPP-AL-NCYW-----------PLNDYGFYTTTGIGYQPYRVVVLS
AAP13567.1 DISNVPFSPDGKPCTPP-AL-NCYW-----------PLNDYGFYTTTGIGYQPYRVVVLS
QHD43416.1 DISTEIYQAGSTPCNGVEGF-NCYF-----------PLQSYGFQPTNGVGYQPYRVVVLS
AVP78031.1 DLSSDE---------------NGVR-----------TLSTYDFNPNVPLEYQATRVVVLS
ABD75323.1 DLSSEE---------------NGVR-----------TLSTYDFNQNVPLEYQATRVVVLS
QDF43835.1 DLTSDE---------------NGVR-----------TLSTYDFYPNVPIEYQATRVVVLS
ABD75332.1 DLSSDE---------------NGVY-----------TLSTYDFYPSIPVEYQATRVVVLS
QDF43820.1 DLSSDDG--------------NGVY-----------TLSTYDFNPNVPVAYQATRVVVLS
AAZ67052.1 DLSSDE---------------NGVR-----------TLSTYDFYPSVPVAYQATRVVVLS
AFS88936.1 EVPQLVNANQYSPCVSI-VP-STVWEDGDYYRKQLSPLEGGGWLVASGSTVAMTEQLQMG
YP_0010399 DVETPLYINPGEYSICRDFSPGGFSEDGQVFKRTLTQFEGGGLLIGVGTRVPMTDNLQMS
:: . :. . : :.
QDF43825.1 FELL----NAPATVC-----GPKLSTDLIKNQCVNFNFNGLTGTGVLTPSSKRFQPFQQF
AGZ48818.1 FELL----NAPATVC-----GPKLSTDLIKNQCVNFNFNGLTGTGVLTPSSKRFQPFQQF
ALK02457.1 FELL----NAPATVC-----GPKLSTDLIKNQCVNFNFNGLTGTGVLTPSSKRFQPFQQF
AAS10463.1 FELL----NAPATVC-----GPKLSTDLIKNQCVNFNFNGLTGTGVLTPSSKRFQPFQQF
AAP13441.1 FELL----NAPATVC-----GPKLSTDLIKNQCVNFNFNGLTGTGVLTPSSKRFQPFQQF
AAP13567.1 FELL----NAPATVC-----GPKLSTDLIKNQCVNFNFNGLTGTGVLTPSSKRFQPFQQF
QHD43416.1 FELL----HAPATVC-----GPKKSTNLVKNKCVNFNFNGLTGTGVLTESNKKFLPFQQF
AVP78031.1 FELL----NAPATVC-----GPKLSTQLVKNQCVNFNFNGLKGTGVLTDSSKRFQSFQQF
ABD75323.1 FELL----NAPATVC-----GPKLSTSLVKNQCVNFNFNGFKGTGVLTDSSKTFQSFQQF
QDF43835.1 FELL----NAPATVC-----GPKLSTGLVKNQCVNFNFNGLRGTGVLTDSSKRFQSFQQF
ABD75332.1 FELL----NAPATVC-----GPKLSTQLVKNQCVNFNFNGLRGTGVLTTSSKRFQSFQQF
QDF43820.1 FELL----NAPATVC-----GPKLSTQLVKNQCVNFNFNGLKGTGVLTDSSKRFQSFQQF
AAZ67052.1 FELL----NAPATVC-----GPKLSTQLVKNQCVNFNFNGLKGTGVLTESSKRFQSFQQF
AFS88936.1 FGITVQYGTDTNSVCPKLEFANDTKIASQLGNCVEYSLYGVSGRGVFQNCTAVGVRQQRF
YP_0010399 FIISVQYGTGTDSVCPMLDLGDSLTITNRLGKCVDYSLYGVTGRGVFQNCTAVGVKQQRF
* : . :** . . . .:**::.: *. * **: .. *.*
QDF43825.1 GRDVSD-FTDSVRDPKTSEILDISPCSFGGVSVITPGTNTSSEVAVLYQDVNCTDVPVAI
AGZ48818.1 GRDVSD-FTDSVRDPKTSEILDISPCSFGGVSVITPGTNTSSEVAVLYQDVNCTDVPVAI
ALK02457.1 GRDVLD-FTDSVRDPKTSEILDISPCSFGGVSVITPGTNTSSEVAVLYQDVNCTDVPVAI
AAS10463.1 GRDVSD-FTDSVRDPKTSEILDISPCSFGGVSVITPGTNASSEVAVLYQDVNCTDVSTLI
AAP13441.1 GRDVSD-FTDSVRDPKTSEILDISPCSFGGVSVITPGTNASSEVAVLYQDVNCTDVSTAI
AAP13567.1 GRDVSD-FTDSVRDPKTSEILDISPCSFGGVSVITPGTNASSEVAVLYQDVNCTDVSTAI
QHD43416.1 GRDIAD-TTDAVRDPQTLEILDITPCSFGGVSVITPGTNTSNQVAVLYQDVNCTEVPVAI
AVP78031.1 GKDASD-FIDSVRDPQTLEILDITPCSFGGVSVITPGTNTSLEVAVLYQDVNCTDVPTTI
ABD75323.1 GRDASD-FTDSVRDPQTLRILDISPCSFGGVSVITPGTNTSSAVAVLYQDVNCTDVPRTI
QDF43835.1 GRDTSD-FTDSVRDPQTLEILDITPCSFGGVSVITPGTNASSEVAVLYQDVNCTDVPTAI
ABD75332.1 GRDTSD-FTDSVRDPQTLEILDISPCSFGGVSVITPGTNASSEVAVLYQDVNCTDVPTSI
QDF43820.1 GRDTSD-FTDSVRDPQTLEILDITPCSFGGVSVITPGTNASSEVAVLYQDVNCTDVPTAI
AAZ67052.1 GRDTSD-FTDSVRDPQTLEILDISPCSFGGVSVITPGTNASSEVAVLYQDVNCTDVPAAI
AFS88936.1 VYDAYQNLVGYYSDDGNYYCLR--ACVSVPVSVIY--DKETKTHATLFGSVACEHISSTM
YP_0010399 VYDSFDNLVGYYSDDGNYYCVR--PCVSVPVSVIY--DKSTNLHATLFGSVACEHVTTMM
* : . * . : .* **** : : *.*: .* * :. :
QDF43825.1 --HADQLTPAWRIYSTGNNVFQTQAGCLIGAEHVDTSY---ECDIPIGAGICASYHTVSS
AGZ48818.1 --HADQLTPSWRVYSTGNNVFQTQAGCLIGAEHVDTSY---ECDIPIGAGICASYHTVSS
ALK02457.1 --HADQLTPSWRVYSTGNNVFQTQAGCLIGAEHVDTSY---ECDIPIGAGICASYHTVSS
AAS10463.1 --HAEQLTPAWRIYSTGNNVFQTQAGCLIGAEHVDTSY---ECDIPIGAGICASYHTVSS
AAP13441.1 --HADQLTPAWRIYSTGNNVFQTQAGCLIGAEHVDTSY---ECDIPIGAGICASYHTVSL
AAP13567.1 --HADQLTPAWRIYSTGNNVFQTQAGCLIGAEHVDTSY---ECDIPIGAGICASYHTVSL
QHD43416.1 --HADQLTPTWRVYSTGSNVFQTRAGCLIGAEHVNNSY---ECDIPIGAGICASYQTQTN
AVP78031.1 --HADQLTPAWRIYATGTNVFQTQAGCLIGAEHVNASY---ECDIPIGAGICASYHTASI
ABD75323.1 --QADQLAPSWRVYTTGPYVFQTQAGCLIGAEHVNASY---QCDIPIGAGICASYHTASH
QDF43835.1 --RADQLTPAWRVYSTGINVFQTQAGCLIGAEHVNASY---ECDIPIGAGICASYHTAST
ABD75332.1 --HADQLTPAWRVYSTGVNVFQTQAGCLIGAEHVNASY---ECDIPIGAGICASYHTASV
QDF43820.1 --RADQLTPAWRVYSTGVNVFQTQAGCLIGAEHVNASY---ECDIPIGAGICASYHTAST
AAZ67052.1 --HADQLTPAWRVYSTGTNVFQTQAGCLIGAEHVNASY---ECDIPIGAGICASYHTAST
AFS88936.1 SQYSRSTRSMLKRRDSTYGPLQTPVGCVLGL--VNSSLFVEDCKLPLGQSLCALPDTPST
YP_0010399 S-QFSRLTQSNLRRRDSNIPLQTAVGCVIGLS--NNSLVVSDCKLPLGQSLCAV-PPVST
:** .**::* : * :*.:*:* .:** . :
QDF43825.1 ----LRSTS----QKSI--------VAYTMSLGADSSIAYSNNTIAIPTNFSISITTEVM
AGZ48818.1 ----LRSTS----QKSI--------VAYTMSLGADSSIAYSNNTIAIPTNFSISITTEVM
ALK02457.1 ----LRSTS----QKSI--------VAYTMSLGADSSIAYSNNTIAIPTNFSISITTEVM
AAS10463.1 ----LRSTS----QKSI--------VAYTMSLGADSSIAYSNNTIAIPTNFSISITTEVM
AAP13441.1 ----LRSTS----QKSI--------VAYTMSLGADSSIAYSNNTIAIPTNFSISITTEVM
AAP13567.1 ----LRSTS----QKSI--------VAYTMSLGADSSIAYSNNTIAIPTNFSISITTEVM
QHD43416.1 SPRRARSVA----SQSI--------IAYTMSLGAENSVAYSNNSIAIPTNFTISVTTEIL
AVP78031.1 ----LRSTS----QKAI--------VAYTMSLGAENSIAYANNSIAIPTNFSISVTTEVM
ABD75323.1 ----LRSTG----QKSI--------VAYTMSLGAENSVAYANNSIAIPTNFSISVTTEVM
QDF43835.1 ----LRSVG----QKSI--------VAYTMSLGAENSIAYANNSIAIPTNFSISVTTEVM
ABD75332.1 ----LRSTG----QKSI--------VAYTMSLGAENSIAYANNSIAIPTNFSISVTTEVM
QDF43820.1 ----LRSVG----QKSI--------VAYTMSLGAENSIAYANNSIAIPTNFSISVTTEVM
AAZ67052.1 ----LRSVG----QKSI--------VAYTMSLGAENSIAYANNSIAIPTNFSISVTTEVM
AFS88936.1 ----LTPRS----VRSVPGEMRLASIAFNHPIQVDQ-LNSSYFKLSIPTNFSFGVTQEYI
YP_0010399 ----FRSYSASQFQLAV--------LNYTSPIVV-TPINSSGFTAAIPTNFSFSVTQEYI
. . :: : :. .: . : : . :*****::.:* * :
QDF43825.1 PVSMAKTSVDCNMYICGDSTECANLLLQYGSFCTQLNRALSGIAVEQDRNTREVFAQVKQ
AGZ48818.1 PVSMAKTSVDCNMYICGDSTECANLLLQYGSFCTQLNRALSGIAVEQDRNTREVFAQVKQ
ALK02457.1 PVSMAKTSVDCNMYICGDSTECANLLLQYGSFCTQLNRALSGIAVEQDRNTREVFAQVKQ
AAS10463.1 PVSMAKTSVDCNMYICGDSTECANLLLQYGSFCRQLNRALSGIAAEQDRNTREVFVQVKQ
AAP13441.1 PVSMAKTSVDCNMYICGDSTECANLLLQYGSFCTQLNRALSGIAAEQDRNTREVFAQVKQ
AAP13567.1 PVSMAKTSVDCNMYICGDSTECANLLLQYGSFCTQLNRALSGIAAEQDRNTREVFAQVKQ
QHD43416.1 PVSMTKTSVDCTMYICGDSTECSNLLLQYGSFCTQLNRALTGIAVEQDKNTQEVFAQVKQ
AVP78031.1 PVSMAKTSVDCTMYICGDSIECSNLLLQYGSFCTQLNRALSGIAIEQDKNTQEVFAQVKQ
ABD75323.1 PVSMAKTSVDCTMYICGDSLECSNLLLQYGSFCTQLNRALSGIAVEQDKNTQEVFAQVKQ
QDF43835.1 PVSMSKTSVDCTMYICGDSQECSNLLLQYGSFCTQLNRALTGIAIEQDKNTQEVFAQVKQ
ABD75332.1 PVSIAKTSVDCTMYICGDSLECSNLLLQYGSFCTQLNRALTGIAIEQDKNTQEVFAQVKQ
QDF43820.1 PVSMAKTSVDCTMYICGDSQECSNLLLQYGSFCTQLNRALTGVALEQDKNTQEVFAQVKQ
AAZ67052.1 PVSMAKTSVDCTMYICGDSLECSNLLLQYGSFCTQLNRALSGIAIEQDKNTQEVFAQVKQ
AFS88936.1 QTTIQKVTVDCKQYVCNGFQKCEQLLREYGQFCSKINQALHGANLRQDDSVRNLFASVKS
YP_0010399 ETSIQKVTVDCKQYVCNGFTRCEKLLVEYGQFCSKINQALHGANLRQDESVYSLYSNIKT
.:: *.:***. *:*.. * :** :**.** ::*.** * ** .. .:: .:*
QDF43825.1 MYKTPTLKD-FGG-FNFSQILPDPLKPTKRSF---IEDLLFNKVTLADAGFMKQYGECL-
AGZ48818.1 MYKTPTLKD-FGG-FNFSQILPDPLKPTKRSF---IEDLLFNKVTLADAGFMKQYGECL-
ALK02457.1 MYKTPTLKD-FGG-FNFSQILPDPLKPTKRSF---IEDLLFNKVTLADAGFMKQYGECL-
AAS10463.1 MYKTPTLKD-FGG-FNFSQILPDPLKPTKRSF---IEDLLFNKVTLADAGFMKQYGECL-
AAP13441.1 MYKTPTLKY-FGG-FNFSQILPDPLKPTKRSF---IEDLLFNKVTLADAGFMKQYGECL-
AAP13567.1 MYKTPTLKY-FGG-FNFSQILPDPLKPTKRSF---IEDLLFNKVTLADAGFMKQYGECL-
QHD43416.1 IYKTPPIKD-FGG-FNFSQILPDPSKPSKRSF---IEDLLFNKVTLADAGFIKQYGDCL-
AVP78031.1 IYKTPPIKD-FGG-FNFSQILPDPSKPSKRSF---IEDLLFNKVTLADAGFIKQYGDCL-
ABD75323.1 MYKTPTIRD-FGG-FNFSQILPDPLKPTKRSF---IEDLLYNKVTLADAGFMKQYADCL-
QDF43835.1 MYKTPAIKD-FGG-FNFSQILPDPSKPTKRSF---IEDLLFNKVTLADAGFMKQYGECL-
ABD75332.1 MYKTPAIKD-FGG-FNFSQILPDPSKPTKRSF---IEDLLFNKVTLADAGFMKQYGECL-
QDF43820.1 MYKTPAIKD-FGG-FNFSQILPDPSKPTKRSF---IEDLLFNKVTLADAGFMKQYGECL-
AAZ67052.1 MYKTPAIKD-FGG-FNFSQILPDPSKPTKRSF---IEDLLFNKVTLADAGFMKQYGECL-
AFS88936.1 SQSSPIIPG-FGGDFNLTLLEPVSISTGSRSARSAIEDLLFDKVTIADPGYMQGYDDCMQ
YP_0010399 T-STQTLEYGLNGDFNLTLLQVPQIGGSSSSYRSAIEDLLFDKVTIADPGYMQGYDDCMK
.: : :.* **:: : . * *****::***:**.*::: * :*:
QDF43825.1 -GDINARDLICAQKFNGLTVLPPLLTDDMIAAYTAALVSGTATAGWTFGAGAALQIPFAM
AGZ48818.1 -GDINARDLICAQKFNGLTVLPPLLTDDMIAAYTAALVSGTATAGWTFGAGAALQIPFAM
ALK02457.1 -GDINARDLICAQKFNGLTVLPPLLTDDMIAAYTAALVSGTATAGWTFGAGAALQIPFAM
AAS10463.1 -GDINARDLICAQKFNGLTVLPPLLTDDMIAAYTAALVSGTATAGWTFGAGAALQIPFAM
AAP13441.1 -GDINARDLICAQKFNGLTVLPPLLTDDMIAAYTAALVSGTATAGWTFGAGAALQIPFAM
AAP13567.1 -GDINARDLICAQKFNGLTVLPPLLTDDMIAAYTAALVSGTATAGWTFGAGAALQIPFAM
QHD43416.1 -GDIAARDLICAQKFNGLTVLPPLLTDEMIAQYTSALLAGTITSGWTFGAGAALQIPFAM
AVP78031.1 -GGISARDLICAQKFNGLTVLPPLLTDEMIAAYTAALISGTATAGWTFGAGAALQIPFAM
ABD75323.1 -GGINARDLICAQKFNGLTVLPPLLTDDMIAAYTAALISGTATAGWTFGAGAALQIPFAM
QDF43835.1 -GDINARDLICAQKFNGLTVLPPLLTDDMIAAYTAALVSGTATAGWTFGAGAALQIPFAM
ABD75332.1 -GDISARDLICAQKFNGLTVLPPLLTDEMIAAYTAALVSGTATAGWTFGAGSALQIPFAM
QDF43820.1 -GDINARDLICAQKFNGLTVLPPLLTDDMIAAYTAALVSGTATAGWTFGAGAALQIPFAM
AAZ67052.1 -GDISARDLICAQKFNGLTVLPPLLTDEMIAAYTAALVSGTATAGWTFGAGSALQIPFAM
AFS88936.1 QGPASARDLICAQYVAGYKVLPPLMDVNMEAAYTSSLLGSIAGVGWTAGLSSFAAIPFAQ
YP_0010399 QGPQSARDLICAQYVSGYKVLPPLYDPNMEAAYTSSLLGSIAGAGWTAGLSSFAAIPFAQ
* ******** . * .***** :* * **::*:.. *** * .: ****
QDF43825.1 QMAYRFNGIGVTQNVLYENQKQIANQFNKAISQIQESLTTTSTALGKLQDVVNQNAQALN
AGZ48818.1 QMAYRFNGIGVTQNVLYENQKQIANQFNKAISQIQESLTTTSTALGKLQDVVNQNAQALN
ALK02457.1 QMAYRFNGIGVTQNVLYENQKQIANQFNKAISQIQESLTTTSTALGKLQDVVNQNAQALN
AAS10463.1 QMAYRFNGIGVTQNVLYENQKQIANQFNKAISQIQESLTTTSTALGKLQDVVNQNAQALN
AAP13441.1 QMAYRFNGIGVTQNVLYENQKQIANQFNKAISQIQESLTTTSTALGKLQDVVNQNAQALN
AAP13567.1 QMAYRFNGIGVTQNVLYENQKQIANQFNKAISQIQESLTTTSTALGKLQDVVNQNAQALN
QHD43416.1 QMAYRFNGIGVTQNVLYENQKLIANQFNSAIGKIQDSLSSTASALGKLQDVVNQNAQALN
AVP78031.1 QMAYRFNGIGVTQNVLYENQKLIANQFNSAIGKIQESLTSTASALGKLQDVVNQNAQALN
ABD75323.1 QMAYRFNGIGVTQNVLYENQKQIANQFNKAITQIQESLTTTSTALGKLQDVVNQNAQALN
QDF43835.1 QMAYRFNGIGVTQNVLYENQKQIANQFNKAISQIQESLTTTSTALGKLQDVVNQNAQALN
ABD75332.1 QMAYRFNGIGVTQNVLYENQKQIANQFNKAISQIQESLTTTSTALGKLQDVVNQNAQALN
QDF43820.1 QMAYRFNGIGVTQNVLYENQKQIANQFNKAISQIQESLTTTSTALGKLQDVVNQNAQALN
AAZ67052.1 QMAYRFNGIGVTQNVLYENQKQIANQFNKAISQIQESLTTTSTALGKLQDVVNQNAQALN
AFS88936.1 SIFYRLNGVGITQQVLSENQKLIANKFNQALGAMQTGFTTTNEAFQKVQDAVNNNAQALS
YP_0010399 SMFYRLNGVGITQQVLSENQKLIANKFNQALGAMQTGFTTSNLAFSKVQDAVNANAQALS
.: **:**:*:**:** **** ***:**.*: :* .:::: *: *:**.** *****.
QDF43825.1 TLVKQLSSNFGAISSVLNDILSRLDKVEAEVQIDRLITGRLQSLQTYVTQQLIRAAEIRA
AGZ48818.1 TLVKQLSSNFGAISSVLNDILSRLDKVEAEVQIDRLITGRLQSLQTYVTQQLIRAAEIRA
ALK02457.1 TLVKQLSSNFGAISSVLNDILSRLDKVEAEVQIDRLITGRLQSLQTYVTQQLIRAAEIRA
AAS10463.1 TLVKQLSSNFGAISSVLNDILSRLDKVEAEVQIDRLITGRLQSLQTYVTQQLIRAAEIRA
AAP13441.1 TLVKQLSSNFGAISSVLNDILSRLDKVEAEVQIDRLITGRLQSLQTYVTQQLIRAAEIRA
AAP13567.1 TLVKQLSSNFGAISSVLNDILSRLDKVEAEVQIDRLITGRLQSLQTYVTQQLIRAAEIRA
QHD43416.1 TLVKQLSSNFGAISSVLNDILSRLDKVEAEVQIDRLITGRLQSLQTYVTQQLIRAAEIRA
AVP78031.1 TLVKQLSSNFGAISSVLNDILSRLDKVEAEVQIDRLITGRLQSLQTYVTQQLIRAAEIRA
ABD75323.1 TLVKQLSSNFGAISSALNDILSRLDKVEAEVQIDRLITGRLQSLQTYVTQQLIRAAEIRA
QDF43835.1 TLVKQLSSNFGAISSVLNDILSRLDKVEAEVQIDRLITGRLQSLQTYVTQQLIRAAEIRA
ABD75332.1 TLVKQLSSNFGAISSVLNDILSRLDKVEAEVQIDRLITGRLQSLQTYVTQQLIRAAEIRA
QDF43820.1 TLVKQLSSNFGAISSVLNDILSRLDKVEAEVQIDRLITGRLQSLQTYVTQQLIRAAEIRA
AAZ67052.1 TLVKQLSSNFGAISSVLNDILSRLDKVEAEVQIDRLITGRLQSLQTYVTQQLIRAAEIRA
AFS88936.1 KLASELSNTFGAISASIGDIIQRLDVLEQDAQIDRLINGRLTTLNAFVAQQLVRSESAAL
YP_0010399 KLASELSNTFGAISSSISDILARLDTVEQDAQIDRLINGRLISLNAFVSQQLVRSETAAR
.*..:**..*****: :.**: *** :* :.******.*** :*:::*:***:*:
QDF43825.1 SANLAATKMSECVLGQSKRVDFCGKGYHLMSFPQAAPHGVVFLHVTYVPSQERNFTTAPA
AGZ48818.1 SANLAATKMSECVLGQSKRVDFCGKGYHLMSFPQAAPHGVVFLHVTYVPSQERNFTTAPA
ALK02457.1 SANLAATKMSECVLGQSKRVDFCGKGYHLMSFPQAAPHGVVFLHVTYVPSQERNFTTAPA
AAS10463.1 SANLAATKMSECVLGQSKRVDFCGKGYHLMSFPQAAPHGVVFLHVTYVPSQERNFTTAPA
AAP13441.1 SANLAATKMSECVLGQSKRVDFCGKGYHLMSFPQAAPHGVVFLHVTYVPSQERNFTTAPA
AAP13567.1 SANLAATKMSECVLGQSKRVDFCGKGYHLMSFPQAAPHGVVFLHVTYVPSQERNFTTAPA
QHD43416.1 SANLAATKMSECVLGQSKRVDFCGKGYHLMSFPQSAPHGVVFLHVTYVPAQEKNFTTAPA
AVP78031.1 SANLAATKMSECVLGQSKRVDFCGKGYHLMSFPQSAPHGVVFLHVTYIPSQEKNFTTAPA
ABD75323.1 SANLAATKMSECVLGQSKRVDFCGKGYHLMSFPQSAPHGVVFLHVTYVPSQEKNFTTAPA
QDF43835.1 SANLAATKMSECVLGQSKRVDFCGRGYHLMSFPQAAPHGVVFLHVTYVPSQEKNFTTAPA
ABD75332.1 SANLAATKMSECVLGQSKRVDFCGKGYHLMSFPQAAPHGVVFLHVTYVPSQERNFTTAPA
QDF43820.1 SANLAATKMSECVLGQSKRVDFCGRGYHLMSFPQAAPHGVVFLHVTYVPSQEKNFTTAPA
AAZ67052.1 SANLAATKMSECVLGQSKRVDFCGKGYHLMSFPQAAPHGVVFLHVTYVPSQERNFTTAPA
AFS88936.1 SAQLAKDKVNECVKAQSKRSGFCGQGTHIVSFVVNAPNGLYFMHVGYYPSNHIEVVSAYG
YP_0010399 SAQLASDKVNECVKSQSKRNGFCGSGTHIVSFVVNAPNGFYFFHVGYVPTNYTNVTAAYG
**:** *:.*** .**** .*** * *::** **:*. *:** * *:: :..:* .
QDF43825.1 ICHEGK---AYFPREGVFVFNGTS-------WFITQRNFFSPQIITTDNT-FVSGSCDVV
AGZ48818.1 ICHEGK---AYFPREGVFVFNGTS-------WFITQRNFFSPQIITTDNT-FVSGSCDVV
ALK02457.1 ICHEGK---AYFPREGVFVFNGTS-------WFITQRNFFSPQIITTDNT-FVSGSCDVV
AAS10463.1 ICHEGK---AYFPREGVFVFNGTS-------WFITQRNFFSPQIITTDNT-FVSGNCDVV
AAP13441.1 ICHEGK---AYFPREGVFVFNGTS-------WFITQRNFFSPQIITTDNT-FVSGNCDVV
AAP13567.1 ICHEGK---AYFPREGVFVFNGTS-------WFITQRNFFSPQIITTDNT-FVSGNCDVV
QHD43416.1 ICHDGK---AHFPREGVFVSNGTH-------WFVTQRNFYEPQIITTDNT-FVSGNCDVV
AVP78031.1 ICHEGK---AHFPREGVFVSNGTH-------WFVTQRNFYEPKIITTDNT-FVSGNCDVV
ABD75323.1 ICHEGK---AYFPREGVFVSNGSS-------WFITQRNFYSPQIITTDNT-FVAGSCDVV
QDF43835.1 ICHEGK---AYFPREGVFVSNGTS-------WFITQRNFYSPQIITTDNT-FVAGSCDVV
ABD75332.1 ICHEGK---AYFPREGVFVSNGTS-------WFITQRNFYSPQIITTDNT-FVAGNCDVV
QDF43820.1 ICHEGK---AYFPREGVFVSNGTF-------WFITQRNFYSPQIITTDNT-FVAGNCDVV
AAZ67052.1 ICHEGK---AYFPREGVFVSNGTS-------WFITQRNFYSPQIITTDNT-FVAGSCDVV
AFS88936.1 LCDAANPTNCIAPVNGYFIKTNNT--RIVDEWSYTGSSFYAPEPITSLNTKYVA--PQVT
YP_0010399 LCNNNNPPLCIAPIDGYFITNQTTTYSVDTEWYYTGSSFYKPEPITQANSRYVS--SDVK
:* : . * :* *: . . * * .*: *: ** *: :*: :*
QDF43825.1 IGIINNTVYDPL---QPELDSFKEELDKYFKNHTSPDVDLGDISGINASVVNIQKEIDRL
AGZ48818.1 IGIINNTVYDPL---QPELDSFKEELDKYFKNHTSPDVDLGDISGINASVVNIQKEINRL
ALK02457.1 IGIINNTVYDPL---QPELDSFKEELDKYFKNHTSPDVDLGDISGINASVVNIQKEIDRL
AAS10463.1 IGIINNTVYDPL---QPELDSFKEELDKYFKNHTSPDVDLGDISGINASVVNIQEEIDRL
AAP13441.1 IGIINNTVYDPL---QPELDSFKEELDKYFKNHTSPDVDLGDISGINASVVNIQKEIDRL
AAP13567.1 IGIINNTVYDPL---QPELDSFKEELDKYFKNHTSPDVDLGDISGINASVVNIQKEIDRL
QHD43416.1 IGIVNNTVYDPL---QPELDSFKEELDKYFKNHTSPDVDLGDISGINASVVNIQKEIDRL
AVP78031.1 IGIINNTVYDPL---QPELDSFKEELDKYFKNHTSPDIDLGDISGINASVVNIQKEIDRL
ABD75323.1 IGIINNTVYDPL---QPELDSFKQELDKYFKNHTSPDVDLGDISGINASVVDIQKEIDRL
QDF43835.1 IGIINNTVYDPL---QPELDSFKEELDKYFKNHTSPDVDLGDISGINASVVNIQKEIDRL
ABD75332.1 IGIINNTVYDPL---QPELDSFKEELDKYFKNHTSPDVDLGDISGINASVVNIQKEIDRL
QDF43820.1 IGIINNTVYDPL---QPELDSFKEELDKYFKNHTSPDVDLGDISGINASVVNIQKEIDRL
AAZ67052.1 IGIINNTVYDPL---QPELDSFKEELDKYFKNHTSPDVDLGDISGINASVVNIQKEIDRL
AFS88936.1 YQNISTNLPPPLLGNSTGID-FQDELDEFFKNVSTSIPNFGSLTQINTTLLDLTYEMLSL
YP_0010399 FDKLENNLPPPLLENSTDVD-FKDELEEFFKNVTSHGPNFAEISKINTTLLDLSDEMAML
:...: ** .. :* *::**:::*** :: ::..:: **:::::: *: *
QDF43825.1 NEVAKNLNESLIDLQELGKYEQYIKWPWYVWLGFIAGLIAIVMVTILLCCMTSCCSCLKG
AGZ48818.1 NEVAKNLNESLIDLQELGKYEQYIKWPWYVWLGFIAGLIAIVMVTILLCCMTSCCSCLKG
ALK02457.1 NEVAKNLNESLIDLQELGKYEQYIKWPWYVWLGFIAGLIAIVMVTILLCCMTSCCSCLKG
AAS10463.1 NEVAKNLNESLIDLQELGKYEQYIKWPWYVWLGFIAGLIAIVMVTILLCCMTSCCSCLKG
AAP13441.1 NEVAKNLNESLIDLQELGKYEQYIKWPWYVWLGFIAGLIAIVMVTILLCCMTSCCSCLKG
AAP13567.1 NEVAKNLNESLIDLQELGKYEQYIKWPWYVWLGFIAGLIAIVMVTILLCCMTSCCSCLKG
QHD43416.1 NEVAKNLNESLIDLQELGKYEQYIKWPWYIWLGFIAGLIAIVMVTIMLCCMTSCCSCLKG
AVP78031.1 NEVARNLNESLIDLQELGKYEQYIKWPWYVWLGFIAGLIAIVMVTILLCCMTSCCSCLKG
ABD75323.1 NEVAKNLNESLIDLQELGKYEQYIKWPWYVWLGFIAGLVGLFMAIILLCYFTSCCSCCKG
QDF43835.1 NEVAKNLNESLIDLQELGKYEQYIKWPWYVWLGFIAGLIAIVMATILLCCMTSCCSCLKG
ABD75332.1 NEVAKNLNESLIDLQELGKYEQYIKWPWYVWLGFIAGLIAIVMVTILLCCMTSCCSCLKG
QDF43820.1 NEVAKNLNESLIDLQELGKYEQYIKWPWYVWLGFIAGLIAIVMATILLCCMTSCCSCLKG
AAZ67052.1 NEVAKNLNESLIDLQELGKYEQYIKWPWYVWLGFIAGLIAIVMVTILLCCMTSCCSCLKG
AFS88936.1 QQVVKALNESYIDLKELGNYTYYNKWPWYIWLGFIAGLVALALCVFFILCCTGCGTNCMG
YP_0010399 QEVVKQLNDSYIDLKELGNYTYYNKWPWYVWLGFIAGLVALLLCVFFLLCCTGCGTSCLG
::*.. **:* ***:***:* * *****:********:.: : ::: *.* : *
QDF43825.1 ACSCGSCC-KFDEDDSEPVLKGVKLHYT
AGZ48818.1 ACSCGSCC-KFDEDDSEPVLKGVKLHYT
ALK02457.1 ACSCGSCC-KFDEDDSEPVLKGVKLHYT
AAS10463.1 ACSCGSCC-KFDEDDSEPVLKGVKLHYT
AAP13441.1 ACSCGSCC-KFDEDDSEPVLKGVKLHYT
AAP13567.1 ACSCGSCC-KFDEDDSEPVLKGVKLHYT
QHD43416.1 CCSCGSCC-KFDEDDSEPVLKGVKLHYT
AVP78031.1 CCSCGSCC-KFDEDDSEPVLKGVKLHYT
ABD75323.1 MCSCGSCC-RFDEDDSEPVLKGVKLHYT
QDF43835.1 ACSCGSCC-KFDEDDSEPVLKGVKLHYT
ABD75332.1 ACSCGSCC-KFDEDDSEPVLKGVKLHYT
QDF43820.1 ACSCGSCC-KFDEDDSEPVLKGVKLHYT
AAZ67052.1 ACSCGSCC-KFDEDDSEPVLKGVKLHYT
AFS88936.1 KLKCNRCCDRYEEYDLEP----HKVHVH
YP_0010399 KMKCKNCCDSYEEYDVE------KIHVH
.* ** ::* * * *:*
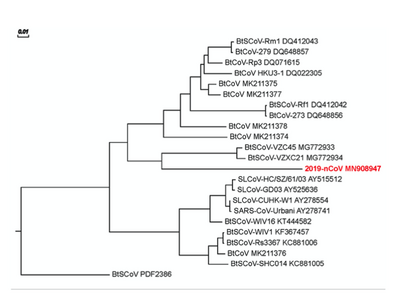
- I then went back and clicked on the Tree Rendering tab, which then showed a phylogenetic tree of the 15 sequences.
- On this tree, horizontal lines (branches) represented individual evolutionary lineages. By contrast, vertical lines (splits) represented mutation events, and the vertical length of each split was drawn purely for visual clarity with no biological meaning. The left-most split was called the root of the tree, and represented a hypothesis about the most recent common ancestor (MRCA) of the sequences within my tree.
- In Figure 2 of Wan et al. (2020), an outgroup called BtSCoV PDF2386 was used. However, Dr. Dahlquist was unable to find this sequence in GenBank for us to use.
- The tree is related heavily to the multiple sequence alignment. Sequences which were found to be very similar are closer to each on the tree and vice versa. For example, AFS88936.1 & YP_0010399 were often very explicit outliers in the sequence alignment, and this is the case in the phylogenetic tree as well. The branch distance between these sequences and the rest reflects the differences in their sequences.
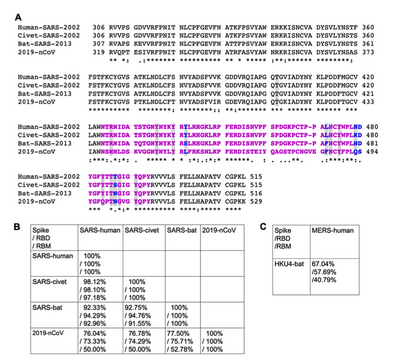
- I compared my alignment to figure 3 from Wan et. al. (2020), pictured above.
- Figure 3 of Wan et al. compares base pairs ~306-515 of the spike protein sequences of Human SARS 2002, civet SARS 2002, bat SARS 2013, and human SARS-CoV-2. Whereas my alignment compares the entire spike protein sequences of 15 (of the 16) viruses they compared in their phylogenetic tree. Below I have narrowed my alignment down to just include those from figure 3.
QDF43825.1 RVAPSKEVVRFPNITNLCPFGEVFNATTF
AGZ48818.1 RVAPSKEVVRFPNITNLCPFGEVFNATTF
ALK02457.1 RVAPSKEVVRFPNITNLCPFGEVFNATTF
AAS10463.1 RVVPSGDVVRFPNITNLCPFGEVFNATKF
AAP13441.1 RVVPSGDVVRFPNITNLCPFGEVFNATKF
AAP13567.1 RVVPSGDVVRFPNITNLCPFGEVFNATKF
QHD43416.1 RVQPTESIVRFPNITNLCPFGEVFNATRF
AVP78031.1 RVQPTQSVVRFPNITNVCPFHKVFNATRF
ABD75323.1 RVSPVTEVVRFPNITNLCPFDKVFNATRF
QDF43835.1 RVSPTHEVIRFPNITNRCPFDKVFNASRF
ABD75332.1 RVTPTQEVVRFPNITNRCPFDKVFNASRF
QDF43820.1 RVSPTQEVVRFPNITNRCPFDKVFNASRF
AAZ67052.1 RVSPTQEVIRFPNITNRCPFDKVFNATRF
AFS88936.1 EAKPSGSVVEQAEGVE-CDFSPLLSGTP-
YP_0010399 EASATGTFIEQPNATE-CDFSPMLTGVA-
. . .: .: .: * * ::..
QDF43825.1 PSVYAWERKRISNCVADYSVLYNSTSFSTFKCYGVSATKLNDLCFSNVYADSFVVKGDDV
AGZ48818.1 PSVYAWERKRISNCVADYSVLYNSTSFSTFKCYGVSATKLNDLCFSNVYADSFVVKGDDV
ALK02457.1 PSVYAWERKRISNCVADYSVLYNSTSFSTFKCYGVSATKLNDLCFSNVYADSFVVKGDDV
AAS10463.1 PSVYAWERKRISNCVADYSVLYNSTSFSTFKCYGVSATKLNDLCFSNVYADSFVVKGDDV
AAP13441.1 PSVYAWERKKISNCVADYSVLYNSTFFSTFKCYGVSATKLNDLCFSNVYADSFVVKGDDV
AAP13567.1 PSVYAWERKKISNCVADYSVLYNSTFFSTFKCYGVSATKLNDLCFSNVYADSFVVKGDDV
QHD43416.1 ASVYAWNRKRISNCVADYSVLYNSASFSTFKCYGVSPTKLNDLCFTNVYADSFVIRGDEV
AVP78031.1 PSVYAWERTKISDCIADYTVFYNSTSFSTFKCYGVSPSKLIDLCFTSVYADTFLIRFSEV
ABD75323.1 PSVYAWERTKISDCVADYTVFYNSTSFSTFNCYGVSPSKLIDLCFTSVYADTFLIRFSEV
QDF43835.1 PNVYAWERTKISDCVADYTVLYNSTSFSTFKCYGVSPSKLIDLCFTSVYADTFLIRSSEV
ABD75332.1 PNVYAWERTKISDCVADYTVLYNSTSFSTFKCYGVSPSKLIDLCFTSVYADTFLIRSSEV
QDF43820.1 PNVYAWERTKISDCVADYTVLYNSTSFSTFKCYGVSPSKLIDLCFTSVYADTFLIRSSEV
AAZ67052.1 PNVYAWERTKISDCVADYTVLYNSTSFSTFKCYGVSPSKLIDLCFTSVYADTFLIRSSEV
AFS88936.1 PQVYNFKRLVFTNCNYNLTKLLSLFSVNDFTCSQISPAAIASNCYSSLILDYFSYPLSMK
YP_0010399 PQVYNFKRLVFSNCNYNLTKLLSLFAVDEFSCNGISPDSIARGCYSTLTVDYFAYPLSMK
..** ::* :::* : : : . .. *.* :*. : *::.: * * .
QDF43825.1 RQIAPGQTGVIADYNYKLPDDFMGC-VLAWNTRNIDATSTGNYNYKYRSLRHGKLRPFER
AGZ48818.1 RQIAPGQTGVIADYNYKLPDDFTGC-VLAWNTRNIDATQTGNYNYKYRSLRHGKLRPFER
ALK02457.1 RQIAPGQTGVIADYNYKLPDDFTGC-VLAWNTRNIDATQTGNYNYKYRSLRHGKLRPFER
AAS10463.1 RQIAPGQTGVIADYNYKLPDDFMGC-VLAWNTRNIDATSTGNYNYKYRYLRHGKLRPFER
AAP13441.1 RQIAPGQTGVIADYNYKLPDDFMGC-VLAWNTRNIDATSTGNYNYKYRYLRHGKLRPFER
AAP13567.1 RQIAPGQTGVIADYNYKLPDDFMGC-VLAWNTRNIDATSTGNYNYKYRYLRHGKLRPFER
QHD43416.1 RQIAPGQTGKIADYNYKLPDDFTGC-VIAWNSNNLDSKVGGNYNYLYRLFRKSNLKPFER
AVP78031.1 RQVAPGQTGVIADYNYKLPDDFTGC-VIAWNTAKQD---VGNYF--YRSHRSTKLKPFER
ABD75323.1 RQVAPGQTGVIADYNYKLPDDFTGC-VIAWNTAKQD---VGSYF--YRSHRSSKLKPFER
QDF43835.1 RQVAPGETGVIADYNYKLPDDFTGC-VIAWNTAKQD---QGQYY--YRSSRKTKLKPFER
ABD75332.1 RQVAPGETGVIADYNYKLPDDFTGC-VIAWNTAQQD---QGQYY--YRSYRKEKLKPFER
QDF43820.1 RQVAPGETGVIADYNYKLPDDFTGC-VIAWNTAKQD---TGHYY--YRSHRKTKLKPFER
AAZ67052.1 RQVAPGETGVIADYNYKLPDDFTGC-VIAWNTAKQD---QGQYY--YRSHRKTKLKPFER
AFS88936.1 SDLSVSSAGPISQFNYKQSFSNPTC-LILATVPHNLTTITKPLKYSYINKCSRLLSDDRT
YP_0010399 SYIRPGSAGNIPLYNYKQSFANPTCRVMASVLANVTITKPHAYG--YIS-KCSRLTGANQ
: ..:* *. :*** . * :: : * *
QDF43825.1 DISNVPFSPDGKPCTPP-AF-NCYW-----------PLNDYGFFTTNGIGYQPYRVVVLS
AGZ48818.1 DISNVPFSPDGKPCTPP-AF-NCYW-----------PLNDYGFYITNGIGYQPYRVVVLS
ALK02457.1 DISNVPFSPDGKPCTPP-AF-NCYW-----------PLNDYGFYITNGIGYQPYRVVVLS
AAS10463.1 DISNVPFSPDGKPCTPP-AP-NCYW-----------PLNGYGFYTTSGIGYQPYRVVVLS
AAP13441.1 DISNVPFSPDGKPCTPP-AL-NCYW-----------PLNDYGFYTTTGIGYQPYRVVVLS
AAP13567.1 DISNVPFSPDGKPCTPP-AL-NCYW-----------PLNDYGFYTTTGIGYQPYRVVVLS
QHD43416.1 DISTEIYQAGSTPCNGVEGF-NCYF-----------PLQSYGFQPTNGVGYQPYRVVVLS
AVP78031.1 DLSSDE---------------NGVR-----------TLSTYDFNPNVPLEYQATRVVVLS
ABD75323.1 DLSSEE---------------NGVR-----------TLSTYDFNQNVPLEYQATRVVVLS
QDF43835.1 DLTSDE---------------NGVR-----------TLSTYDFYPNVPIEYQATRVVVLS
ABD75332.1 DLSSDE---------------NGVY-----------TLSTYDFYPSIPVEYQATRVVVLS
QDF43820.1 DLSSDDG--------------NGVY-----------TLSTYDFNPNVPVAYQATRVVVLS
AAZ67052.1 DLSSDE---------------NGVR-----------TLSTYDFYPSVPVAYQATRVVVLS
AFS88936.1 EVPQLVNANQYSPCVSI-VP-STVWEDGDYYRKQLSPLEGGGWLVASGSTVAMTEQLQMG
YP_0010399 DVETPLYINPGEYSICRDFSPGGFSEDGQVFKRTLTQFEGGGLLIGVGTRVPMTDNLQMS
:: . :. . : :.
QDF43825.1 FELL----NAPATVC-----GPKL
AGZ48818.1 FELL----NAPATVC-----GPKL
ALK02457.1 FELL----NAPATVC-----GPKL
AAS10463.1 FELL----NAPATVC-----GPKL
AAP13441.1 FELL----NAPATVC-----GPKL
AAP13567.1 FELL----NAPATVC-----GPKL
QHD43416.1 FELL----HAPATVC-----GPKK
AVP78031.1 FELL----NAPATVC-----GPKL
ABD75323.1 FELL----NAPATVC-----GPKL
QDF43835.1 FELL----NAPATVC-----GPKL
ABD75332.1 FELL----NAPATVC-----GPKL
QDF43820.1 FELL----NAPATVC-----GPKL
AAZ67052.1 FELL----NAPATVC-----GPKL
AFS88936.1 FGITVQYGTDTNSVCPKLEFANDT
YP_0010399 FIISVQYGTGTDSVCPMLDLGDSL
* : . :** . .
- note that:
- “*” indicates invariant
- “:” indicates highly conserved
- “.” indicates weakly conserved
- a space indicates not conserved
- It can be seen that there is much less invariance in my alignment, which is to be expected when comparing many more sequences. Figure 3 also compares the % of sequence similarity among all combinations of sequences (3B), and the sequence similarities of MERS-CoV and HKU4 viral spike proteins. I also noticed that there are many more spaces in the sequences of my alignment, which is necessary when aligning that many varying sequences.
- I then compared my tree to Figure 2 of the Wan et al. (2020) paper, shown above.
- As previously noted, I was unable to incorporate an outgroup called BtSCoV PDF2386 from the Wan et al. paper into my own tree. As a result my tree's outgroup comprises Human betacoronavirus 2c (AFS88936.1) and Trylonycteris bat coronavirus (YP_0010399). My tree was also exclusively based on the spike proteins of these viruses, whereas Wan et al. compared several more sequences for each virus. This is most likely why the trees do not seem to branch in similar patterns.
- I think that there is enough information provided within the Wan et al. (2020) paper to reproduce their analysis, only if you are already familiar with the programs they used/named. I had no idea how to reproduce their trees/alignments until reading Dr. Dahlquist's directions, but after doing this I can see how someone with more experience could intuit their procedures.
Scientific Conclusion
Reproducing some of the findings of the Wan et al. paper was a viable method of gaining experience obtaining sequence data, comparing it using multiple sequence alignments, and analyzing it for phylogenetic relationships using trees. I can continue to expose myself to these programs in order to eventually be able to use them to answer my own questions.
Acknowledgments
- I consulted with my partner Anna Horvath during class and over text to troubleshoot minor technical issues.
- I copied and modified procedures from the week 4 assignment page
- I linked to Nathan Beshai's uploads of the Wan et al. figures from week 3.
- The Notepad desktop application was used to ensure that my sequences were in plain text.
- Phylogeny.fr was used to generate my phylogenetic tree and sequence alignments.
- Except for what is noted above, this individual journal entry was completed by me and not copied from another source.
Aiden Burnett (talk) 20:21, 29 September 2020 (PDT)
References
- Wan, Y., Shang, J., Graham, R., Baric, R. S., & Li, F. (2020). Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. Journal of virology, 94(7). DOI: 10.1128/JVI.00127-20
- OpenWetWare. (2020). BIOL368/F20:Week 4. Retrieved September 27, 2020, from https://openwetware.org/wiki/BIOL368/F20:Week_4
- Bat SARS-like coronavirus Rs3367, complete genome - Nucleotide - NCBI. (2013, November 22). Retrieved September 30, 2020, from https://www.ncbi.nlm.nih.gov/nuccore/556015127/
- Phylogeny.fr: "One Click" Mode. (2020). Retrieved 29 September 2020, from http://www.phylogeny.fr/simple_phylogeny.cgi?workflow_id=b9c0813cbbe9695d63cf7e31da5f026d&tab_index=1
User Page
Template
Course Homepage
Weekly Assignments
- Week 1 Assignment
- Week 2 Assignment
- Week 3 Assignment
- Week 4 Assignment
- Week 5 Assignment
- Week 6 Assignment
- Week 7 Assignment
- Week 8 Assignment
- Week 9 Assignment
- Week 10 Assignment
- Week 11 Assignment
- Week 12 Assignment
- Week 14 Assignment
Individual Journal Pages
- Aiden Burnett
- Aiden Burnett Week 2
- Aiden Burnett Week 3
- Aiden Burnett Week 4
- Aiden Burnett Week 5
- Aiden Burnett Week 6
- Aiden Burnett Week 7
- FoldamerDB Review
- Aiden Burnett Week 9
- Aiden Burnett Week 10
- Aiden Burnett Week 11
- Aiden Burnett Week 12
- Aiden Burnett Week 14