Aiden Burnett Week 6
Purpose
To discern, through sequence comparison, which likely intermediate host for SARS-CoV-2 has the most similar ACE2 receptor to humans. This could then inform comparisons of coronaviruses in future studies by pointing to ones originating from the host(s) we identify,
Methods & Results
- My partner, Anna Horvath , and I first gathered list of possible intermediate hosts from Wan et al, animals present at Wuhan wt markets, and other species. These were:
- humans [1]
- civet [2]
- Chinese bats [3]
- mice [4]
- rats [5]
- pigs [6]
- ferrets [7]
- cats [8]
- orangutans [9]
- grivet monkeys [10]
- fox [11]
- chickens [12]
- king cobras [13]
- We wanted to use the Chinese cobra, as these are present at Wuhan wet markets, but the ACE2 receptor sequence for this speccies was notavailable. We elected to use the king cobra as an alternative.
- pangolins [14]
- dromedary camels [15]
- squirrels [16]
- mink [17]
- Chinese softshell turtles [18]
- We then amassed the ACE2 sequences for these species using UnitProt & Genbank.
- partial sequences were only able to be found for the mink, and so this animal was removed in order to prevent disruptions of sequence analyses.
- a phylogenetic tree was made comparing all of the gathered sequences (excluding mink).
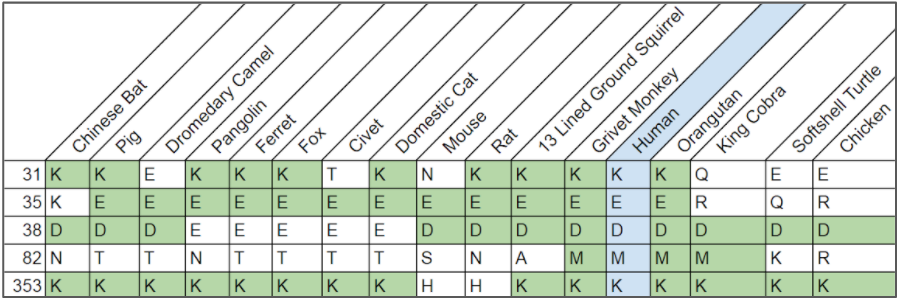
- critical residues of the civet ACE2 were found from the Wan et all paper. These were:
- T31
- E35
- E38
- T82
- K353
- My partner, Anna Horvath, performed the sequence alignment using phylogeny.fr & generated the following phylogenetic tree.
These were used to compare critical residues of every species in the sequence alignment and generate the following table comparing critical residues to
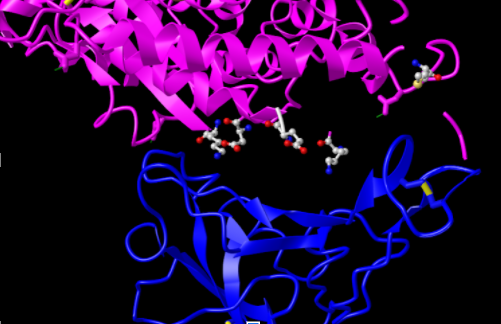
- The critical residues of the human ACE2 were visualized by clicking on the link below to interact with the structure in iCn3D
- I clicked on the Analysis menu to “View Sequences & Annotations”
- In the new window that appeared to the right, I clicked on the “Details” tab to show the actual amino acid sequences
- Focused on the pink chain and oriented it to resemble Figure 4B of Wan et al.
- Next I endeavored to make the amino acid side chains shown in the figure visible.
- In the sequence window I went to sequence “Protein 7A91_D” (in pink) and selected the following amino acids
- K31
- E35
- D38
- M82
- K353
- Went to the Styles menu and selected Proteins > Ball and Stick
- Went to the Color menu and selected Atom
- This allowed me to see the side chains as shown in the figure.
- I was unable to repeat this process for the coronavirus spike protein as I could not identify the corresponding critical residues but the image still illustrates how the residues illustrated might interact with the spike protein
- I then took the following screenshot, oriented like Figure 4B of Wan et al. (2020)
- Lastly I created a presentation based on the information presented above.
Scientific Conclusion
Orangutans and grivet monkeys are have the most similar ACE2 receptors to humans of the possible intermediate hosts. This makes sense as they are known orthologues to humans. Of the five critical amino acids that correspond to the RBD of SARS-CoV-2 on the ACE2 receptor, many were relatively conserved across species, with most organisms having between 2-3 of the 5 amino acids altered. This could inform future research aimed at investigating possible lineages of the SARS-CoV-2 virus. As a result of this research, such studies could include coronaviruses which infect orangutans and grivet monkeys into structure/residue comparisons and phylogenetic trees.
Acknowledgments
- I consulted with my partner Anna Horvath during class and over text to formulate our research question and procedures, and coordinate work on the project.
- my partner Anna Horvath created the phylogenetic tree of ACE2 sequences using phylogeny.fr
- I copied and modified procedures from the week 5 assignment page.
- I used the Microsoft Excel and Google sheets programs in making the residue comparison table.
- Except for what is noted above, this individual journal entry was completed by me and not copied from another source.
Aiden Burnett (talk) 23:30, 14 October 2020 (PDT)
References
- angiotensin-converting enzyme 2 isoform 1 precursor [Homo sapiens] - Protein - NCBI. (2020). Retrieved 8 October 2020, from https://www.ncbi.nlm.nih.gov/protein/NP_001358344.1?report=fasta
- angiotensin-converting enzyme 2 [Paguma larvata] - Protein - NCBI. (2020). Retrieved 8 October 2020, from https://www.ncbi.nlm.nih.gov/protein/AAX63775.1?report=fasta
- angiotensin-converting enzyme 2 [Rhinolophus sinicus] - Protein - NCBI. (2020). Retrieved 8 October 2020, from https://www.ncbi.nlm.nih.gov/protein/AGZ48803.1?report=fasta
- angiotensin-converting enzyme 2 precursor [Mus musculus] - Protein - NCBI. (2020). Retrieved 8 October 2020, from https://www.ncbi.nlm.nih.gov/protein/NP_001123985.1?report=fasta
- angiotensin converting enzyme 2 [Rattus norvegicus] - Protein - NCBI. (2020). Retrieved 8 October 2020, from https://www.ncbi.nlm.nih.gov/protein/AAW78017.1?report=fasta
- angiotensin-converting enzyme 2 precursor [Sus scrofa] - Protein - NCBI. (2020). Retrieved 8 October 2020, from https://www.ncbi.nlm.nih.gov/protein/NP_001116542.1?report=fasta
- angiotensin I converting enzyme 2 [Mustela putorius furo] - Protein - NCBI. (2020). Retrieved 8 October 2020, from https://www.ncbi.nlm.nih.gov/protein/BAE53380.1?report=fasta
- angiotensin I converting enzyme 2 [Felis catus] - Protein - NCBI. (2020). Retrieved 8 October 2020, from https://www.ncbi.nlm.nih.gov/protein/AAX59005.1?report=fasta
- angiotensin-converting enzyme 2 precursor [Pongo abelii] - Protein - NCBI. (2020). Retrieved 8 October 2020, from https://www.ncbi.nlm.nih.gov/protein/NP_001124604.1?report=fasta
- angiotensin converting enzyme 2 [Chlorocebus aethiops] - Protein - NCBI. (2020). Retrieved 8 October 2020, from https://www.ncbi.nlm.nih.gov/protein/AAY57872.1?report=fasta
- angiotensin-converting enzyme 2 [Vulpes vulpes] - Protein - NCBI. (2020). Retrieved 8 October 2020, from https://www.ncbi.nlm.nih.gov/protein/XP_025842513.1?report=fasta
- angiotensin-converting enzyme 2 [Gallus gallus] - Protein - NCBI. (2020). Retrieved 8 October 2020, from https://www.ncbi.nlm.nih.gov/protein/XP_416822.2?report=fasta
- Angiotensin-converting enzyme 2 [Ophiophagus hannah] - Protein - NCBI. (2020). Retrieved 8 October 2020, from https://www.ncbi.nlm.nih.gov/protein/ETE61880.1?report=fasta
- Angiotensin I converting enzyme 2 [Manis pentadactyla] - Protein - NCBI. (n.d.). Retrieved October 08, 2020, from https://www.ncbi.nlm.nih.gov/protein/QLH93383.1?report=fasta
- Angiotensin-converting enzyme 2 [Camelus dromedarius] - Protein - NCBI. (n.d.). Retrieved October 08, 2020, from https://www.ncbi.nlm.nih.gov/protein/XP_031301717.1
- Angiotensin-converting enzyme 2, partial [Neovison vison] - Protein - NCBI. (2020). Retrieved 8 October 2020, from https://www.ncbi.nlm.nih.gov/protein/CCP86723.1?report=fasta
- angiotensin-converting enzyme 2 [Ictidomys tridecemlineatus] - Protein - NCBI. (2020). Retrieved 8 October 2020, from https://www.ncbi.nlm.nih.gov/protein/XP_005316051.3?report=fasta
- angiotensin-converting enzyme 2 [Pelodiscus sinensis] - Protein - NCBI. (2020). Retrieved 8 October 2020, from https://www.ncbi.nlm.nih.gov/protein/XP_006122891.1?report=fasta
- Wan, Y., Shang, J., Graham, R., Baric, R., & Li, F. (2020). Receptor Recognition by the Novel Coronavirus from Wuhan: an Analysis Based
on Decade-Long Structural Studies of SARS Coronavirus. Journal Of Virology, 94(7). doi: 10.1128/jvi.00127-20
Yuan, S., Jiang, S., & Li, Z. (2020). Analysis of Possible Intermediate Hosts of the New Coronavirus SARS-CoV-2. Frontiers In Veterinary Science, 7. doi: 10.3389/fvets.2020.00379
- OpenWetWare. (2020). BIOL368/F20:Week 5. Retrieved October 5, 2020, from https://openwetware.org/wiki/BIOL368/F20:Week_5
User Page
Template
Course Homepage
Weekly Assignments
- Week 1 Assignment
- Week 2 Assignment
- Week 3 Assignment
- Week 4 Assignment
- Week 5 Assignment
- Week 6 Assignment
- Week 7 Assignment
- Week 8 Assignment
- Week 9 Assignment
- Week 10 Assignment
- Week 11 Assignment
- Week 12 Assignment
- Week 14 Assignment
Individual Journal Pages
- Aiden Burnett
- Aiden Burnett Week 2
- Aiden Burnett Week 3
- Aiden Burnett Week 4
- Aiden Burnett Week 5
- Aiden Burnett Week 6
- Aiden Burnett Week 7
- FoldamerDB Review
- Aiden Burnett Week 9
- Aiden Burnett Week 10
- Aiden Burnett Week 11
- Aiden Burnett Week 12
- Aiden Burnett Week 14