Yaniv Maddahi Journal Week 5
From OpenWetWare
Jump to navigationJump to search
Purpose
- The purpose of this lab is to develop our skills of structure building programs and to investigate and modify the relationship and structure of the Spike Protein RBD and ACE2. We will be using the programs to analyze the difference aspects of the Spike and ACE2 interactions and to represent them in various ways. We will be learning to manipulate the structure in order to understand and view different aspects of their interactions. We will also postulate our own research question for our own future studying.
Methods/Results
- I found out what is already known about the spike protein in the UniProt Knowledgebase (UniProt KB). UniProt KB has two parts to it, Swis-Prot, which contains entries for proteins that have been manually reviewed, and TrEMBL (which stands for "Translated EMBL"), which are automated translations of all DNA sequences in the EMBL/GenBank/DDBJ databases. SARS-CoV-2 is so new that it was only added to the database with the April 22 release.
- I searched using the keyword "SARS-CoV-2" in the main UniProt search field and used the entry with accession number "P0DTC2" which corresponds to the reference entry for the SARS-CoV-2 spike protein.
- I viewed one of the structures of the SARS-CoV spike protein from Wan et al. (2020) in the NCBI Structure Database using the web-based iCn3D viewer, I chose figure 2A SARS-CoV RBD (year 2002) complexed with human ACE2: 2AJF
- I created a view of the protein that recreated Figure 2A from the Wan et al. (2020) paper.
- I clicked on the Style > Proteins menu and show screenshots of the following styles:
- Cylinder and Plate
- C Alpha Trace
- Lines
- Ball and Stick
- Spheres
- I then went to the “Spheres” view, clicked on the Color menu and chose the following color schemes:
- Spectrum
- Secondary
- Charge
- Atom
- I found the N-terminus and C-terminus of each polypeptide.
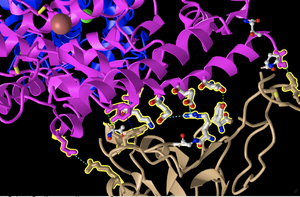
- The looked at side chain-side chain interactions in the civet ACE2-spike protein structure shown in Figure 4B by first clicking on the link to interact with the structure in iCn3D.
- I clicked on the Windows menu to “View Sequences & Annotations”.
- In the new window that appears to the right, I clicked on the “Details” tab to show the actual amino acid sequences. There are 2 sets of ACE2-spike proteins because of the way the proteins crystallized.
- I focused on the pink and tan chains and orient them like is shown in Figure 4B
- I made the amino acid side chains shown in the figure visible.
- In the sequence window, I went to sequence “Protein 3SCK_A” (in pink) and select the following amino acids:
- T31
- E35
- E38
- T82
- K353
- The part of the ribbon that represents these amino acids were highlighted in yellow in the structure
- I went to the Styles menu and selected Proteins > Ball and Stick, went to the Color menu, and selected Atom. Side chains were shown in the figure.
- Repeated this process for the tan spike protein sequence 3SCK_E for the following amino acids:
- T487
- R479
- G480
- Y442
- P472
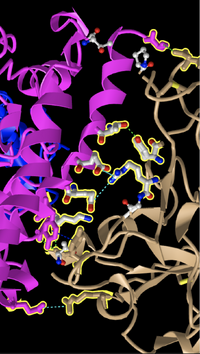
- I made the dashed lines for these bonds in iCn3D as follows:
- I went to the View menu and select H bonds & Interactions
- In part 1 of the window that appears, I unchecked “Contacts/Interactions” leaving Hydrogen Bonds and Ionic Interaction checked
- In part 2 of the window, I selected the first set “3SCK_A” (pink)
- In part 3 of the window, I selected the second set “3SCK_E” (tan)
- In part 4 of the window, I clicked the button “3D Display”
UniProtKB Results
- Upon searching SARS-CoV-2 in the UniProt database, there appear to be a total of 1,616 results, 1,598 being unreviewed results and 18 being reviewed results.
- No, they are not all viral proteins; some appear to be human proteins and some mouse proteins.
- Organisms vary from Human to mouse and there appear to be 1,603 organisms, 1,507 strains, and 1,603 taxonomies for SARS-CoV-2.
Protein Structure
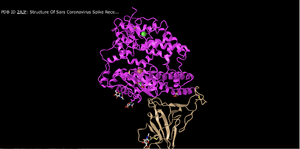
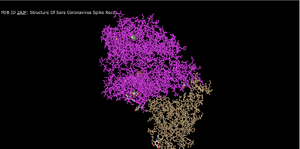
Standard Structure
- This image shows the interactions between ACE2 (Pink) and Spike Protein RBD (Tan).
- This protein structure is of a quaternary structure as there are two separate polypeptide chains interacting with each other via non covalent bonds.
- There are two domains within this structure as the two separate polypeptide chains can be separated without breaking either of the chains.
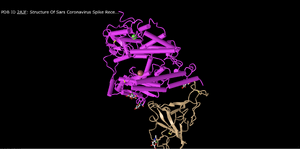
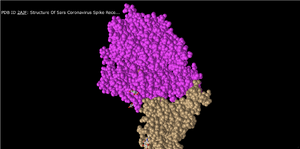
Cylinder and Plate Structure
- In this, the alpha helices appear to be condensed into cylinders and the beta sheets unchanged.
- This makes the overall structure slightly easier to view because the helices to not seem as cluttered.
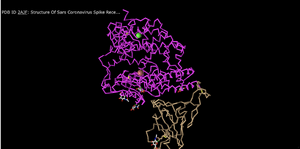
C Alpha Trace
- This seems to give a carbon chain/backbone, omitting the secondary structures.
- This is a very clean style of representation although does not give much information as to the structures that compile the overall protein shape.
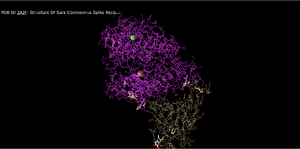
Lines
- This seems to be depicting all of the amino acids and backbones in line structures. It still does omit secondary structures but gives insight to potential backbone interactions.
- This would be useful for observing nuances within the structure but not necessarily for observing the bigger picture.
Ball and Stick
- This is similar to the lines model however here all the atoms are ball instead of lines.
- This is a good representation because it gives more perspective on the structure of how each atom is lined up.
Spheres
- In this setting, each amino acid appears as a cloud/sphere and closely represents what we had previously been taught to call a space filling model.
- This is a good representation because it gives perspective to the amount of space taken up by the atoms in the structure.
Spheres Cont.
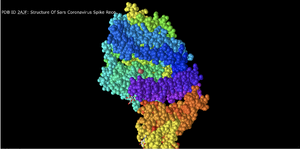
Spheres Spectrum
- This model seems to break the structure into different strata/regions although the method of classification for each region is difficult to discern.
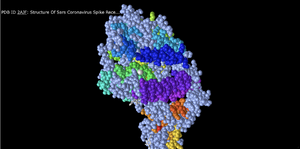
Spheres Secondary
- Using the setting of secondary --> spectrum, the structure above was generated. We believe that perhaps this setting color coordinates the secondary structures within the overall protein structure.
Spheres Charge
- We believe this setting separates amino acids based on category, with acidic amino acids being red, basic amino acids being blue and the light purple, and all uncharged amino acids being gray.
- This is a good visualization because it can give insight into the way in which amino acids and different regions of the protein may interact with each other and the environment.
Spheres Atom
- Here all the atoms are color coordinated by the system in which carbon is gray, nitrogen is red, oxygen is blue, and sulfur is gold/yellow.
- This method of visualization can be especially useful in depicting disulfide bonds if there are any, and help to further predict/ illustrate how the protein may interact with its environment.
- It helps discern the location of atoms from each other and within the protein.
N- and C- Terminus
- As indicated in the figure above.
Secondary Structures of ACE2
- ACE2 contained both alpha helices and beta sheets. The Alpha helices were identified by their coil-like shapes and the Beta sheets by the arrows. There were also random coils identified by being straight lines.
Secondary Structures of Spike Protein
- The Spike protein only contained Beta Sheets and random coils. The Beta sheets are indicated by arrows and the random coils by the straight lines.
E35-R479
- E35 is acidic and R479 is basic. We know this for several reasons; first we know that E represents Glutamic Acid and that Glutamic Acid is acidic. Not only, but we know that Glutamic Acid has a carboxyl group side chain and that Oxygen is depicted in the color red. We also know that Arginine has amino side chains and that Nitrogen is depicted in the color Blue.
T31-Y442
- Both amino acids belong to the "uncharged polar" chemical groups. T represents Threonine and Y represents Tyrosine. Both Amino Acids contain a Hydroxyl (OH) group.
Hydrogen Bonding & Interactions
- See above image for bonding between ACE2 and Spike Protein. Dashed lines indicated H bonds and other non covalent interactions. The Pink structure is ACE2 and the Tan structure is Spike Protein.
Research Question
- How do differences in ACE2 receptors for host organisms affect binding strength with SARS-CoV?
- We will use the UniProt database and possibly GenBank for obtaining sequences. We will remain within the mammals category and possibly look at the sequences for ACE2 of Humans, Bats, Civet Cats, Pangolins, Mice, Squirrels, Rats, Birds, Dogs, Cats, Lions, Tigers, Ferrets, and Minks.
Conclusion
- In this lab we were able to develop our skills at interpreting different models of the ACE2 and Spike Protein. We were able to see different ways to depict both structures and further able to represent the interactions present. We viewed the interactions from different perspectives and further developed our own research question for future study. We were able to use the different softwares available to depict the non covalent interactions between the two structures and identify different regions such as the N and C terminals.
Acknowledgements
- I acknowledge my partner, JT Correy, with whom I collaborated for the protein images, answers to some of the questions, and research question.
- I acknowledge, Kam D. Dahlquist, Ph.D., with whom I met via zoom both in and outside of class regarding questions in completing the assignment.
- I acknowledge, Anna Horvath, whose "reference" I used as a reference for creating my own.
- I acknowledge my TA, Annika G. Dinulos, whom I contacted regarding reviewing my assignment.
- I copied and modified procedures from the Week 5 assignment page.
- I used the Wan et. al article for references in the assignment.
- I explored spike protein model data from UniProt Knowledgebase (UniProt KB).
- I viewed protein models using iCn3D viewer.
- I uploaded images using the Wiki Upload page.
"Except for what is noted above, this individual journal entry was completed by me and not copied from another source." Yaniv Maddahi (talk) 09:52, 6 October 2020 (PDT)
References
- OpenWetWare. (2020). BIOL368/F20:Week 5. Retrieved 1 October 2020, from https://openwetware.org/wiki/BIOL368/F20:Week_5
- OpenWetWare. (2020). BIOL368/F20:Week 1. Retrieved 1 October 2020, from https://openwetware.org/wiki/BIOL368/F20:Week_1
- OpenWetWare. (2020). Anna Horvath Week 5. Retrieved 6 October 2020, https://openwetware.org/wiki/Anna_Horvath_Week_5
- Foldit - Blogs. (2020). Retrieved 6 October 2020, from https://fold.it/portal/blog
- Foldit - Solve Puzzles for Science. (2020). Retrieved 6 October 2020, from https://fold.it/portal/
- iCn3D: Web-based 3D Structure Viewer 2AJF. (2020). Retrieved 6 October 2020, from https://www.ncbi.nlm.nih.gov/Structure/icn3d/full.html?&mmdbid=35213&bu=1&showanno=1
- iCn3D: Web-based 3D Structure Viewer 3SCK. (2020). Retrieved 1 October 2020, from https://www.ncbi.nlm.nih.gov/Structure/icn3d/full.html?pdbid=%203SCK
- Uniprot. (2020). S - Spike glycoprotein precursor - Severe acute respiratory syndrome coronavirus 2 (2019-nCoV) - S gene & protein. Retrieved 1 October 2020, from https://www.uniprot.org/uniprot/P0DTC2
- Wan, Y., Shang, J., Graham, R., Baric, R., & Li, F. (2020). Receptor Recognition by the Novel Coronavirus from Wuhan: an Analysis Based on Decade-Long Structural Studies of SARS Coronavirus. Journal Of Virology, 94(7). doi: 10.1128/jvi.00127-20
- Andersen, K.G., Rambaut, A., Lipkin, W.I. et al. The proximal origin of SARS-CoV-2. Nat Med 26, 450–452 (2020). https://doi.org/10.1038/s41591-020-0820-9
Template
Assignment Week
- Week 1 Assignment
- Week 2 Assignment
- Week 3 Assignment
- Week 4 Assignment
- Week 5 Assignment
- Week 6 Assignment
- Week 7 Assignment
- Week 8 Assignment
- Week 9 Assignment
- Week 10 Assignment
- Week 11 Assignment
- Week 12 Assignment
- Week 14 Assignment
Individual Journal Pages
- Yaniv Maddahi Journal Week 1
- Yaniv Maddahi Journal Week 2
- Yaniv Maddahi Journal Week 3
- Yaniv Maddahi Journal Week 4
- Yaniv Maddahi Journal Week 5
- Yaniv Maddahi Journal Week 6
- Yaniv Maddahi Journal Week 7
- Allosteric Database Review
- Yaniv Maddahi Journal Week 9
- Yaniv Maddahi Journal Week 10
- Yaniv Maddahi Journal Week 11
- The Mutants Research Project Week 12
- The Mutants Research Project Week 14