Week 10 Individual Journal William P Fuchs
From OpenWetWare
Jump to navigationJump to search
Question
- Will there be a visible protein sequence change that correlates with a decline in CD4-T cell count and what are those changes altering the characteristic of the amino acid sequence.
Purpose
- Our purpose was to continue using the programs from BIOL368/F16:Week 9 to better investigate our question and hypothesis.
Methods
- Access Biology Workbench and and upload Subject 10's clone sequences for amino acids and nucleotides and across all visits.
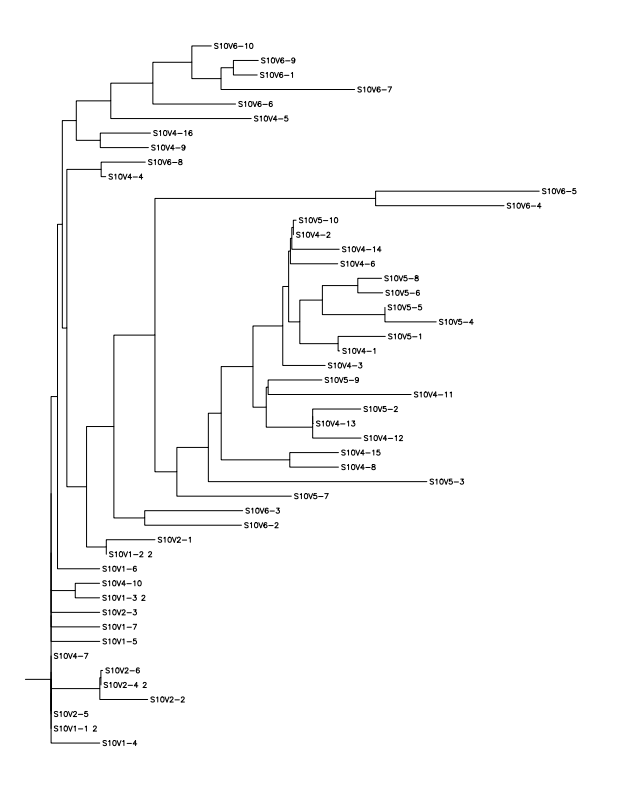
- Run a CLUSTALW multiple sequence alignment and generate a rooted tree.
- Calculate S and Theta
- Collect Min and Max values
- Select the Multiple Sequence alignment tool and run a CLUSTALDIST test on the subject for amino acids.
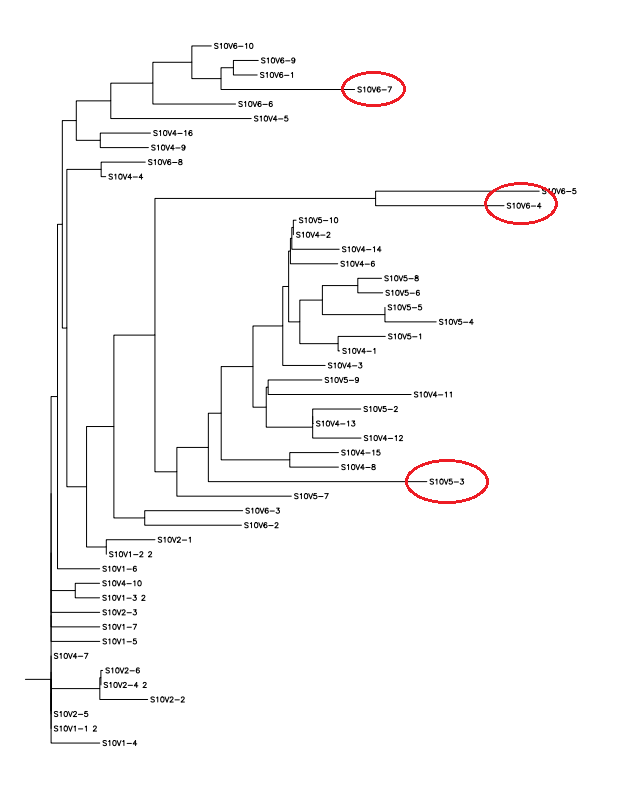
- Find the most divergent visit and clone sequence by resourcing the CLUSTALDIST matrix.
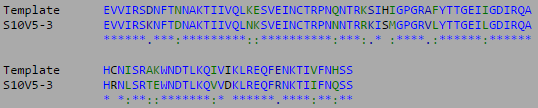
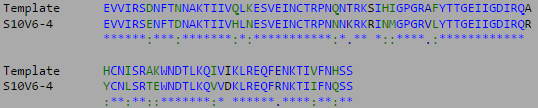
- Isolate the compared amino acid sequences to the V3 target sequence provided by the Huang et al. (2005) data set.
- ...(Continued at Week 11)
Data and Files
Link to Week 10 presentation: here.
Results
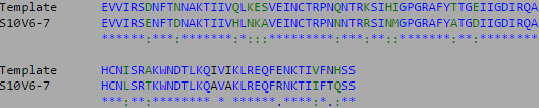
- This week's work toward answering our question yielded interesting results. After preliminary testing of the cultivated clones for their amino acid sequences the clones that showed the highest divergence were data points S10V6-4, S10V5-3, and S10V5-7. Upon analyzing the phylogeny of these constituents since seroconversion they showed the highest divergence in terms of differences in amino acids since initial testings of the subject. Furthermore, there is an observed increase in divergence, since the first visit. Each sets of visits their clones associate in a sequential increase in divergence as further testings were conducted. High amino acid change in samples like S10V6-4, S10V6-5, and S10V6-7 may contribute to the worsening condition associated in the "rapid progressor" category of the Markham et al. (1998) study.
Scientific Conclusion
- Samples S10V6-4, S10V5-3, and S10V5-7 beg the most attention in the continuation of our HIV project and the investigating of the amino acid sequences that describe them.
Acknowledgments
- I worked with Matt Oki in discussing the further directions of the project and the partial presentation of our current work and the results we've obtained thus far. We discussed these matters over the phone and in class
While I worked with the people noted above, this individual journal entry was completed by me and not copied from another source.William P Fuchs 01:58, 7 November 2016 (EST)
References
- Week 10 Assignment Page
- Provided guidelines and instructions for this week's individual journal.
- ExPASY Translate tool
- UniProt Knowledgebase (UniProt KB)
- PredictProtein server
- Cn3D software site
- Huang, C. C., Tang, M., Zhang, M. Y., Majeed, S., Montabana, E., Stanfield, R. L., ... & Wyatt, R. (2005). Structure of a V3-containing HIV-1 gp120 core. Science, 310(5750), 1025-1028. doi: 10.1126/science.1118398.
- Markham, R.B., Wang, W.C., Weisstein, A.E., Wang, Z., Munoz, A., Templeton, A., Margolick, J., Vlahov, D., Quinn, T., Farzadegan, H., & Yu, X.F. (1998). Patterns of HIV-1 evolution in individuals with differing rates of CD4 T cell decline. Proc Natl Acad Sci U S A. 95, 12568-12573.doi: 10.1073/pnas.95.21.12568.
Externals
Individual Journals
- WPF Week 2
- WPF Week 3
- WPF Week 4
- WPF Week 5
- WPF Week 6
- WPF Week 7
- WPF Week 8
- WPF Week 9
- WPF Week 10
- WPF Week 11
- WPF Week 12
- WPF Week 14
- WPF Week 15
Class Journals
- Week 1
- Week 2
- Week 3
- Week 4
- Week 5
- Week 6
- Week 7
- Week 8
- Week 9
- Week 10
- Week 11
- Week 12
- Week 13
- Week 14
- Week 15
Assignments
Useful Links
- Bio Class Page
- BIOL368/F16:People
- Will Fuchs
- Link to LMU: http://www.lmu.edu/