Nyeo2 Week 13
From OpenWetWare
Jump to navigationJump to search
Template
User Page
Assignments
Individual Journals
- Nyeo2 Week 2
- Nyeo2 Week 3
- Nyeo2 Week 4
- Nyeo2 Week 5
- Nyeo2 Week 6
- Nyeo2 Week 8
- Nyeo2 Week 9
- Nyeo2 Week 10
- Nyeo2 Week 11
- Nyeo2 Week 13
- Nyeo2 Week 14
Class Journals
- BIOL368/S20:Class Journal Week 1
- BIOL368/S20:Class Journal Week 2
- BIOL368/S20:Class Journal Week 3
- BIOL368/S20:Class Journal Week 4
- BIOL368/S20:Class Journal Week 5
- BIOL368/S20:Class Journal Week 6
- BIOL368/S20:Bibliography Week 8
- Class Journal Week 9
- BIOL368/S20:Class Journal Week 10
- [[BIOL368/S20:Class Journal Week 11
- BIOL368/S20:Class Journal Week 13
- BIOL368/S20:Class Journal Week 14
Purpose
The purpose of this assignment is to analyze the structure and sequence of the SARS-CoV-2 spike glycoprotein and become familiar with the biological tools to do so.
Methods/Results
Converting spike protein DNA sequence into a protein sequence
- Used spike protein DNA sequence from QHD43416.1: spike protein (Severe acute respiratory syndrome coronavirus 2 isolate Wuhan-Hu-1)
- FASTA DNA Sequence:
>spike protein (Severe acute respiratory syndrome coronavirus 2 isolate Wuhan-Hu-1) DNA sequence ATGTTTGTTTTTCTTGTTTTATTGCCACTAGTCTCTAGTCAGTGTGTTAATCTTACAACCAGAACTCAAT TACCCCCTGCATACACTAATTCTTTCACACGTGGTGTTTATTACCCTGACAAAGTTTTCAGATCCTCAGT TTTACATTCAACTCAGGACTTGTTCTTACCTTTCTTTTCCAATGTTACTTGGTTCCATGCTATACATGTC TCTGGGACCAATGGTACTAAGAGGTTTGATAACCCTGTCCTACCATTTAATGATGGTGTTTATTTTGCTT CCACTGAGAAGTCTAACATAATAAGAGGCTGGATTTTTGGTACTACTTTAGATTCGAAGACCCAGTCCCT ACTTATTGTTAATAACGCTACTAATGTTGTTATTAAAGTCTGTGAATTTCAATTTTGTAATGATCCATTT TTGGGTGTTTATTACCACAAAAACAACAAAAGTTGGATGGAAAGTGAGTTCAGAGTTTATTCTAGTGCGA ATAATTGCACTTTTGAATATGTCTCTCAGCCTTTTCTTATGGACCTTGAAGGAAAACAGGGTAATTTCAA AAATCTTAGGGAATTTGTGTTTAAGAATATTGATGGTTATTTTAAAATATATTCTAAGCACACGCCTATT AATTTAGTGCGTGATCTCCCTCAGGGTTTTTCGGCTTTAGAACCATTGGTAGATTTGCCAATAGGTATTA ACATCACTAGGTTTCAAACTTTACTTGCTTTACATAGAAGTTATTTGACTCCTGGTGATTCTTCTTCAGG TTGGACAGCTGGTGCTGCAGCTTATTATGTGGGTTATCTTCAACCTAGGACTTTTCTATTAAAATATAAT GAAAATGGAACCATTACAGATGCTGTAGACTGTGCACTTGACCCTCTCTCAGAAACAAAGTGTACGTTGA AATCCTTCACTGTAGAAAAAGGAATCTATCAAACTTCTAACTTTAGAGTCCAACCAACAGAATCTATTGT TAGATTTCCTAATATTACAAACTTGTGCCCTTTTGGTGAAGTTTTTAACGCCACCAGATTTGCATCTGTT TATGCTTGGAACAGGAAGAGAATCAGCAACTGTGTTGCTGATTATTCTGTCCTATATAATTCCGCATCAT TTTCCACTTTTAAGTGTTATGGAGTGTCTCCTACTAAATTAAATGATCTCTGCTTTACTAATGTCTATGC AGATTCATTTGTAATTAGAGGTGATGAAGTCAGACAAATCGCTCCAGGGCAAACTGGAAAGATTGCTGAT TATAATTATAAATTACCAGATGATTTTACAGGCTGCGTTATAGCTTGGAATTCTAACAATCTTGATTCTA AGGTTGGTGGTAATTATAATTACCTGTATAGATTGTTTAGGAAGTCTAATCTCAAACCTTTTGAGAGAGA TATTTCAACTGAAATCTATCAGGCCGGTAGCACACCTTGTAATGGTGTTGAAGGTTTTAATTGTTACTTT CCTTTACAATCATATGGTTTCCAACCCACTAATGGTGTTGGTTACCAACCATACAGAGTAGTAGTACTTT CTTTTGAACTTCTACATGCACCAGCAACTGTTTGTGGACCTAAAAAGTCTACTAATTTGGTTAAAAACAA ATGTGTCAATTTCAACTTCAATGGTTTAACAGGCACAGGTGTTCTTACTGAGTCTAACAAAAAGTTTCTG CCTTTCCAACAATTTGGCAGAGACATTGCTGACACTACTGATGCTGTCCGTGATCCACAGACACTTGAGA TTCTTGACATTACACCATGTTCTTTTGGTGGTGTCAGTGTTATAACACCAGGAACAAATACTTCTAACCA GGTTGCTGTTCTTTATCAGGATGTTAACTGCACAGAAGTCCCTGTTGCTATTCATGCAGATCAACTTACT CCTACTTGGCGTGTTTATTCTACAGGTTCTAATGTTTTTCAAACACGTGCAGGCTGTTTAATAGGGGCTG AACATGTCAACAACTCATATGAGTGTGACATACCCATTGGTGCAGGTATATGCGCTAGTTATCAGACTCA GACTAATTCTCCTCGGCGGGCACGTAGTGTAGCTAGTCAATCCATCATTGCCTACACTATGTCACTTGGT GCAGAAAATTCAGTTGCTTACTCTAATAACTCTATTGCCATACCCACAAATTTTACTATTAGTGTTACCA CAGAAATTCTACCAGTGTCTATGACCAAGACATCAGTAGATTGTACAATGTACATTTGTGGTGATTCAAC TGAATGCAGCAATCTTTTGTTGCAATATGGCAGTTTTTGTACACAATTAAACCGTGCTTTAACTGGAATA GCTGTTGAACAAGACAAAAACACCCAAGAAGTTTTTGCACAAGTCAAACAAATTTACAAAACACCACCAA TTAAAGATTTTGGTGGTTTTAATTTTTCACAAATATTACCAGATCCATCAAAACCAAGCAAGAGGTCATT TATTGAAGATCTACTTTTCAACAAAGTGACACTTGCAGATGCTGGCTTCATCAAACAATATGGTGATTGC CTTGGTGATATTGCTGCTAGAGACCTCATTTGTGCACAAAAGTTTAACGGCCTTACTGTTTTGCCACCTT TGCTCACAGATGAAATGATTGCTCAATACACTTCTGCACTGTTAGCGGGTACAATCACTTCTGGTTGGAC CTTTGGTGCAGGTGCTGCATTACAAATACCATTTGCTATGCAAATGGCTTATAGGTTTAATGGTATTGGA GTTACACAGAATGTTCTCTATGAGAACCAAAAATTGATTGCCAACCAATTTAATAGTGCTATTGGCAAAA TTCAAGACTCACTTTCTTCCACAGCAAGTGCACTTGGAAAACTTCAAGATGTGGTCAACCAAAATGCACA AGCTTTAAACACGCTTGTTAAACAACTTAGCTCCAATTTTGGTGCAATTTCAAGTGTTTTAAATGATATC CTTTCACGTCTTGACAAAGTTGAGGCTGAAGTGCAAATTGATAGGTTGATCACAGGCAGACTTCAAAGTT TGCAGACATATGTGACTCAACAATTAATTAGAGCTGCAGAAATCAGAGCTTCTGCTAATCTTGCTGCTAC TAAAATGTCAGAGTGTGTACTTGGACAATCAAAAAGAGTTGATTTTTGTGGAAAGGGCTATCATCTTATG TCCTTCCCTCAGTCAGCACCTCATGGTGTAGTCTTCTTGCATGTGACTTATGTCCCTGCACAAGAAAAGA ACTTCACAACTGCTCCTGCCATTTGTCATGATGGAAAAGCACACTTTCCTCGTGAAGGTGTCTTTGTTTC AAATGGCACACACTGGTTTGTAACACAAAGGAATTTTTATGAACCACAAATCATTACTACAGACAACACA TTTGTGTCTGGTAACTGTGATGTTGTAATAGGAATTGTCAACAACACAGTTTATGATCCTTTGCAACCTG AATTAGACTCATTCAAGGAGGAGTTAGATAAATATTTTAAGAATCATACATCACCAGATGTTGATTTAGG TGACATCTCTGGCATTAATGCTTCAGTTGTAAACATTCAAAAAGAAATTGACCGCCTCAATGAGGTTGCC AAGAATTTAAATGAATCTCTCATCGATCTCCAAGAACTTGGAAAGTATGAGCAGTATATAAAATGGCCAT GGTACATTTGGCTAGGTTTTATAGCTGGCTTGATTGCCATAGTAATGGTGACAATTATGCTTTGCTGTAT GACCAGTTGCTGTAGTTGTCTCAAGGGCTGTTGTTCTTGTGGATCCTGCTGCAAATTTGATGAAGACGAC TCTGAGCCAGTGCTCAAAGGAGTCAAATTACATTACACATAA
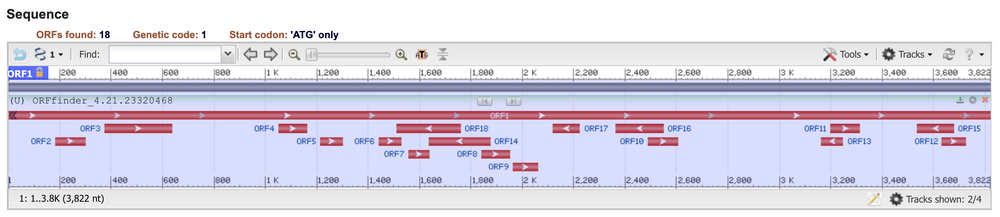
- Copied and pasted sequence into NCBI Open Reading Frame Finder
- Protein sequence:
>lcl|ORF1 MFVFLVLLPLVSSQCVNLTTRTQLPPAYTNSFTRGVYYPDKVFRSSVLHS TQDLFLPFFSNVTWFHAIHVSGTNGTKRFDNPVLPFNDGVYFASTEKSNI IRGWIFGTTLDSKTQSLLIVNNATNVVIKVCEFQFCNDPFLGVYYHKNNK SWMESEFRVYSSANNCTFEYVSQPFLMDLEGKQGNFKNLREFVFKNIDGY FKIYSKHTPINLVRDLPQGFSALEPLVDLPIGINITRFQTLLALHRSYLT PGDSSSGWTAGAAAYYVGYLQPRTFLLKYNENGTITDAVDCALDPLSETK CTLKSFTVEKGIYQTSNFRVQPTESIVRFPNITNLCPFGEVFNATRFASV YAWNRKRISNCVADYSVLYNSASFSTFKCYGVSPTKLNDLCFTNVYADSF VIRGDEVRQIAPGQTGKIADYNYKLPDDFTGCVIAWNSNNLDSKVGGNYN YLYRLFRKSNLKPFERDISTEIYQAGSTPCNGVEGFNCYFPLQSYGFQPT NGVGYQPYRVVVLSFELLHAPATVCGPKKSTNLVKNKCVNFNFNGLTGTG VLTESNKKFLPFQQFGRDIADTTDAVRDPQTLEILDITPCSFGGVSVITP GTNTSNQVAVLYQDVNCTEVPVAIHADQLTPTWRVYSTGSNVFQTRAGCL IGAEHVNNSYECDIPIGAGICASYQTQTNSPRRARSVASQSIIAYTMSLG AENSVAYSNNSIAIPTNFTISVTTEILPVSMTKTSVDCTMYICGDSTECS NLLLQYGSFCTQLNRALTGIAVEQDKNTQEVFAQVKQIYKTPPIKDFGGF NFSQILPDPSKPSKRSFIEDLLFNKVTLADAGFIKQYGDCLGDIAARDLI CAQKFNGLTVLPPLLTDEMIAQYTSALLAGTITSGWTFGAGAALQIPFAM QMAYRFNGIGVTQNVLYENQKLIANQFNSAIGKIQDSLSSTASALGKLQD VVNQNAQALNTLVKQLSSNFGAISSVLNDILSRLDKVEAEVQIDRLITGR LQSLQTYVTQQLIRAAEIRASANLAATKMSECVLGQSKRVDFCGKGYHLM SFPQSAPHGVVFLHVTYVPAQEKNFTTAPAICHDGKAHFPREGVFVSNGT HWFVTQRNFYEPQIITTDNTFVSGNCDVVIGIVNNTVYDPLQPELDSFKE ELDKYFKNHTSPDVDLGDISGINASVVNIQKEIDRLNEVAKNLNESLIDL QELGKYEQYIKWPWYIWLGFIAGLIAIVMVTIMLCCMTSCCSCLKGCCSC GSCCKFDEDDSEPVLKGVKLHYT
- Sequence diagram:
- Also copied and pasted nucleotide sequence in ExPASY Translate tool
- 5'3' Reading frame 1:
- The correct reading frame is reading frame 1, or ORF1 because it runs the span of the entire sequence, goes 5' to 3' and starts with M at the beginning, denoting an AUG start codon.
UniProt KB database search
- Searched for "SARS-CoV" in UniProt Knowledgebase (UniProt KB).
- Received 818 total results (70 reviewed, 748 unreviewed)
- Used the entry with accession number "P59594", which was for the SARS-CoV spike protein.(reviewed)
- Information provided:
- function of each subunit
- molecular functions
- biological processes its involved in
- taxonomy
- virus hosts
- proteomes
- subcellular location
- structural topology
- cellular components
- pathology and biotech
- molecular processing locations
- amino acid modifications
- post-translational modifications
- family and domains
- cleavage sites
- structure visualizations
- protein interactions
- genomic sequence
- similar proteins
- cross-references
- Information provided:
Predicting the SARS-CoV-2 spike protein sequence
- From Walls, A. C., Park, Y. J., Tortorici, M. A., Wall, A., McGuire, A. T., & Veesler, D. (2020). Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. DOI: 10.1016/j.cell.2020.02.058
- 6VXX(spike glycoprotein closed state)
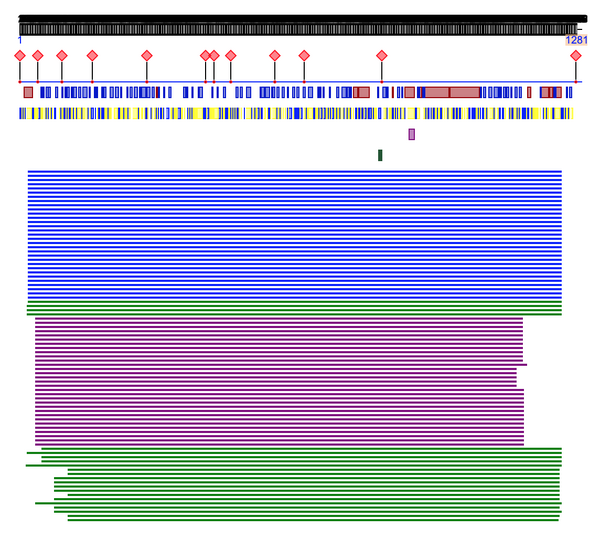
- Pasted FASTA amino acid sequence into PredictProtein server.
- Sequence length: 1281
- 84 aligned proteins and 31 matched PDB structures
- Predicted protein:
- Amino acid composition:
- 6VYB(spike glycoprotein open state)
- Pasted FASTA amino acid sequence into PredictProtein server.
- Sequence length: 1281
- 86 aligned proteins and 31 matched PDB structures
- Predicted protein:

- Amino acid composition:
- 6VXX(spike glycoprotein closed state)
- Structure annotations(left side of page):
- solvent accessibility and secondary structure
- transmembrane helices
- protein disorder and flexibility
- disulphide bridges
- Function annotations(left side of page):
- effect of point mutations
- gene ontology terms
- subcellular localization
- binding sites
- The UniProt database contains information that has been already gathered about a specific protein while the PredictProtein server can predict the result of different structural or functional annotations to the known sequence, such as disulphide bridges or point mutations.
Analyzing the structure of the spike protein
- From Walls et al. (2020): 6VYB
- This is the SARS-CoV-2 spike ectodomain structure (open state)
- In PDB: Under 3D view, clicked "structure"
- Changed view to JSmol
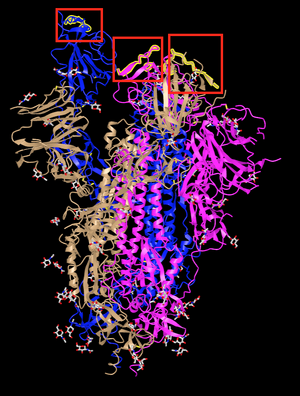
- Figure 3D from Walls et al. (2020)(left) and screenshot from PDB(right)
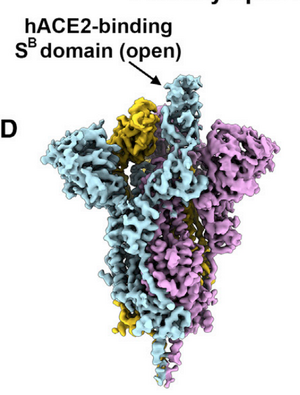
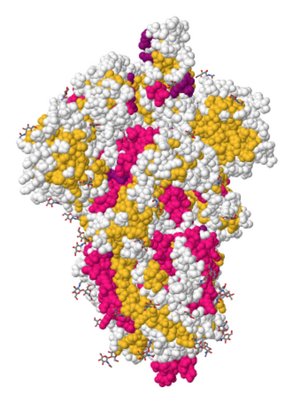
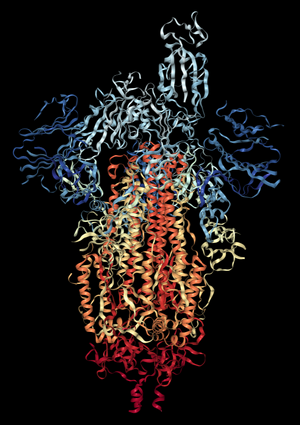
- Under NGL view on PDB - N and C terminus can be seen, denoted as blue and red, respectively
- Black background, cartoon style, rainbow color
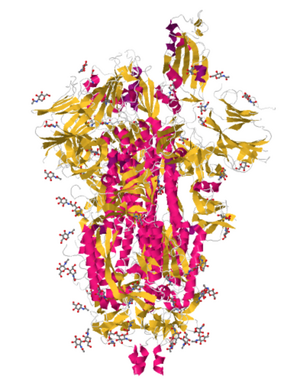
- Under JSmol view on PDB - alpha helices(pink) and beta sheets(yellow)
- Secondary structure color, cartoon style
- The findings do match what was found by the PredictProtein server
- Searched for "6VYB" in NCBI Structure home page and clicked on "full-featured 3D viewer"
- In the upper right corner, searched for the amino acid sequence from positions 486 to 505 in figure 2C in Walls et al. (2020)
- The boxed and highlighted portions are 9 of the 14 key residues for binding hACE2
Data and Files
- sequence diagram
- 5'3' reading frame 1
- 6VXX predicted protein
- 6VXX amino acid composition
- 6VYB predicted protein
- 6VYB amino acid composition
- Walls et al.(2020) Figure 3D
- Protein Database screenshot
- N and C terminus
- secondary structure
- amino acids
Research project
- In Yan et al. (2020), the difference in residues in RBD-PD complex of SARS-CoV and SARS-CoV-2 resulted in weaker molecular interactions between the virus and the receptor. Interactions are important because, as found in Walls et al. (2020), the 2003-04 SARS reemergence was much less deadly than the 2002 outbreak because the virus had a weaker interaction with ACE2, and thus the patients showed milder symptoms. This research project will further explore the role that certain amino acids play in the structure-function relationship of SARS-CoV-2 and ACE2.
- We will analyze sequences from ACE2 sequences from humans, mice, and bats, since it is found or theorized that the virus can infect them.
- The sequences will be taken from UniProt and we will compare them through a multiple sequence alignment. Their 3D structures will also be compared using the NCBI structure function, the protein data bank, or other programs to see if any structural make an impact on the ability of the virus's S protein to interact with the host environment.
Scientific Conclusion
This week we used several tools, such as the NCBI open reading frame finder and the Protein databank to view sequences and structures of the SARS-CoV-2 spike glycoprotein. The data that was found was compared to the findings in the paper from the previous week. By doing this exercise, the we have now read about the function and have visualized the structure of the spike glycoprotein. The goal is to put them both together in a final project.
Acknowledgements
- My homework partners for the week were Drew Cartmel and Jack Menzagopian
- I followed the protocol on BIOL368/S20:Week 13 to complete this assignment
- I also copied the links to the tools and pages from the assignment page
- Followed the wiki syntax on Wikipedia:Manual of Style/Images to format screenshots
- I texted Madeleine King for help on finding the amino acid sequences on the SARS-CoV-2 S structure
- Except for what is noted above, this individual journal entry was completed by me and not copied from another source
Nyeo2 (talk) 20:45, 22 April 2020 (PDT)
References
- ExPASy. (2020). Translate. Retrieved April 22, 2020 from https://web.expasy.org/translate/.
- NCBI. (2020). PDB ID 6VYB: SARS-CoV-2 spike ectodomain structure (open state). Retrieved April 22, 2020 from https://www.ncbi.nlm.nih.gov/Structure/pdb/6VYB
- OpenWetWare. (2020). BIOL368/S20:Week 13. Retrieved April 22, 2020, from https://openwetware.org/wiki/BIOL368/S20:Week_13.
- PredictProtein. (2020). PredictProtein Open. Retrieved April 22, 2020 from https://open.predictprotein.org/.
- RCSB Protein Data Bank. (2020). 6VYB and 6VXY. Retrieved April 22, 2020 from https://www.rcsb.org/structure/6VYB and https://www.rcsb.org/structure/6VXY
- UniProt KB. Retrieved April 22, 2020 from http://www.uniprot.org
- Wikipedia:Manual of Style/Images. Retrieved April 22, 2020 from https://en.wikipedia.org/wiki/Wikipedia:Manual_of_Style/Images#Horizontal_placement