Nyeo2 Week 14
Template
User Page
Assignments
Individual Journals
- Nyeo2 Week 2
- Nyeo2 Week 3
- Nyeo2 Week 4
- Nyeo2 Week 5
- Nyeo2 Week 6
- Nyeo2 Week 8
- Nyeo2 Week 9
- Nyeo2 Week 10
- Nyeo2 Week 11
- Nyeo2 Week 13
- Nyeo2 Week 14
Class Journals
- BIOL368/S20:Class Journal Week 1
- BIOL368/S20:Class Journal Week 2
- BIOL368/S20:Class Journal Week 3
- BIOL368/S20:Class Journal Week 4
- BIOL368/S20:Class Journal Week 5
- BIOL368/S20:Class Journal Week 6
- BIOL368/S20:Bibliography Week 8
- Class Journal Week 9
- BIOL368/S20:Class Journal Week 10
- [[BIOL368/S20:Class Journal Week 11
- BIOL368/S20:Class Journal Week 13
- BIOL368/S20:Class Journal Week 14
Purpose
The SARS-CoV epidemic in 2002 briefly reemergence in 2003-04, but the pathogenicity of the reemerged virus was lower and patients showed milder symptoms. According to Walls et al., 2020, the reemerged virus had weaker interactions with hACE2. SARS-CoV-2, the virus responsible for the current pandemic, shows stronger polar interactions with certain regions of with hACE2 according to Yan et al.,2020. This research project aims to see if the pathogenicity of SARS-CoV and SARS-CoV-2 is related to the strength of the polar interactions between the virus and ACE2 by comparing the different polar residues of the three different viruses.
Methods/Results
Multiple Sequence Alignment
- Aligned sequences(see "Data and Files") on www.phylogeny.fr
- Used "One Click" under "Phylogeny Analysis"
- Uploaded FASTA protein sequences for all S glycoproteins
- Exported the alignment as clustal format
- Aligned Sequence:
AAP50485.1 MFI--------FLLFLTLTS-------GSDLDRCTTFDDVQ--APNYTQHTSSMRGVYYP
AAP30713.1 MFI--------FLLFLTLTS-------GSDLDRCTTFDDVQ--APNYTQHTSSMRGVYYP
AAP13441.1 MFI--------FLLFLTLTS-------GSDLDRCTTFDDVQ--APNYTQHTSSMRGVYYP
AAP41037.1 MFI--------FLLFLTLTS-------GSDLDRCTTFDDVQ--APNYTQHTSSMRGVYYP
AAU04664.1 MFI--------FLLFLTLTS-------GSDLDRCTTFDDVQ--APNYTQHTSSMRGVYYP
AAV49730.1 MFI--------FLLFLTLTS-------GSDLDRCTTFDDVQ--APNYTQHTSSMRGVYYP
AAV97995.1 MFI--------FLLFLTLTS-------GSDLDRCTTFDDVQ--APNYTQHTSSMRGVYYP
AAV98000.1 MFI--------FLLFLTLTS-------GSDLDRCTTFDDVQ--APNYTQHTSSMRGVYYP
n6VXX_1|Ch MGILPSPGMPALLSLVSLLSVLLMGCVAETGTQCVNLTTRTQLPPAYTN--SFTRGVYYP
QHR63300.2 MFV--------FLVLLPLVS-----------SQCVNLTTRTQLPPAYTN--SSTRGVYYP
sp|P0DTC2| MFV--------FLVLLPLVS-----------SQCVNLTTRTQLPPAYTN--SFTRGVYYP
QHD43416.1 MFV--------FLVLLPLVS-----------SQCVNLTTRTQLPPAYTN--SFTRGVYYP
* : :* ::.* * .*..: .* **: * ******
AAP50485.1 DEIFRSDTLYLTQDLFLPFYSNVTGFHTINHT-------FGNPVIPFKDGIYFAATEKSN
AAP30713.1 DEIFRSDTLYLTQDLFLPFYSNVTGFHTINHT-------FGNPVIPFKDGIYFAATEKSN
AAP13441.1 DEIFRSDTLYLTQDLFLPFYSNVTGFHTINHT-------FGNPVIPFKDGIYFAATEKSN
AAP41037.1 DEIFRSDTLYLTQDLFLPFYSNVTGFHTINHT-------FGNPVIPFKDGIYFAATEKSN
AAU04664.1 DEIFRSDTLYLTQDLFLPFYSNVTGFHTINHT-------FDNPVIPFKDGIYFAATEKSN
AAV49730.1 DEIFRSDTLYLTQDLFLPFYSNVTGFHTINHT-------FDNPVIPFKDGIYFAATEKSN
AAV97995.1 DEIFRSDTLYLTQDLFLPFYSNVTGFHTINHT-------FDNPVIPFKDGIYFAATEKSN
AAV98000.1 DEIFRSDTLYLTQDLFLPFYSNVTGFHTINHT-------FDNPVIPFKDGIYFAATEKSN
n6VXX_1|Ch DKVFRSSVLHSTQDLFLPFFSNVTWFHAIHVSGTNGTKRFDNPVLPFNDGVYFASTEKSN
QHR63300.2 DKVFRSSVLHLTQDLFLPFFSNVTWFHAIHVSGTNGIKRFDNPVLPFNDGVYFASTEKSN
sp|P0DTC2| DKVFRSSVLHSTQDLFLPFFSNVTWFHAIHVSGTNGTKRFDNPVLPFNDGVYFASTEKSN
QHD43416.1 DKVFRSSVLHSTQDLFLPFFSNVTWFHAIHVSGTNGTKRFDNPVLPFNDGVYFASTEKSN
*::***..*: ********:**** **:*: : *.***:**:**:***:*****
AAP50485.1 VVRGWVFGSTMNNKSQSVIIINNSTNVVIRACNFELCDNPFFAVSKPMGTQTHT----MI
AAP30713.1 VVRGWVFGSTMNNKSQSVIIINNSTNVVIRACNFELCDNPFFAVSKPMGTQTHT----MI
AAP13441.1 VVRGWVFGSTMNNKSQSVIIINNSTNVVIRACNFELCDNPFFAVSKPMGTQTHT----MI
AAP41037.1 VVRGWVFGSTMNNKSQSVIIINNSTNVVIRACNFELCDNPFFAVSKPMGTQTHT----MI
AAU04664.1 VVRGWVFGSTMNNKSQSVIIINNSTNVVIRACNFELCDNPFFVVSKPMGTRTHT----MI
AAV49730.1 VVRGWVFGSTMNNKSQSVIIINNSTNVVIRACNFELCDNPFFVVSKPMGTRTHT----MI
AAV97995.1 VVRGWVFGSTMNNKSQSVIIINNSTNVVIRACNFELCDNPFFVVSKPMGTQTHT----MI
AAV98000.1 VVRGWVFGSTMNNKSQSVIIINNSTNVVIRACNFELCDNPFFVVSKPMGTQTHT----MI
n6VXX_1|Ch IIRGWIFGTTLDSKTQSLLIVNNATNVVIKVCEFQFCNDPFLGVYYHKNNKSWMESEFRV
QHR63300.2 IIRGWIFGTTLDSKTQSLLIVNNATNVVIKVCEFQFCNDPFLGVYYHKNNKSWMESEFRV
sp|P0DTC2| IIRGWIFGTTLDSKTQSLLIVNNATNVVIKVCEFQFCNDPFLGVYYHKNNKSWMESEFRV
QHD43416.1 IIRGWIFGTTLDSKTQSLLIVNNATNVVIKVCEFQFCNDPFLGVYYHKNNKSWMESEFRV
::***:**:*::.*:**::*:**:*****..*:*::*::**: * ...:. :
AAP50485.1 FDNAFNCTFEYISDAFSLDVSEKSGNFKHLREFVFKNKDGFLYVYKGYQPIDVVRDLPSG
AAP30713.1 FDNAFNCTFEYISDAFSLDVSEKSGNFKHLREFVFKNKDGFLYVYKGYQPIDVVRDLPSG
AAP13441.1 FDNAFNCTFEYISDAFSLDVSEKSGNFKHLREFVFKNKDGFLYVYKGYQPIDVVRDLPSG
AAP41037.1 FDNAFNCTFEYISDAFSLDVSEKSGNFKHLREFVFKNKDGFLYVYKGYQPIDVVRDLPSG
AAU04664.1 FDNAFNCTFEYISDAFSLDVSEKSGNFKHLREFVFKNKDGFLYVYKGYQPIDVVRDLPSG
AAV49730.1 FDNAFNCTFEYISDAFSLDVSEKSGNFKHLREFVFKNKDGFLYVYKGYQPIDVVRDLPSG
AAV97995.1 FDNAFNCTFEYISDAFSLDVSEKSGNFKHLREFVFKNKDGFLYVYKGYQPIDVVRDLPSG
AAV98000.1 FDNAFNCTFEYISDAFSLDVSEKSGNFKHLREFVFKNKDGFLYVYKGYQPIDVVRDLPSG
n6VXX_1|Ch YSSANNCTFEYVSQPFLMDLEGKQGNFKNLREFVFKNIDGYFKIYSKHTPINLVRDLPQG
QHR63300.2 YSSANNCTFEYVSQPFLMDLEGKQGNFKNLREFVFKNIDGYFKIYSKHTPINLVRDLPPG
sp|P0DTC2| YSSANNCTFEYVSQPFLMDLEGKQGNFKNLREFVFKNIDGYFKIYSKHTPINLVRDLPQG
QHD43416.1 YSSANNCTFEYVSQPFLMDLEGKQGNFKNLREFVFKNIDGYFKIYSKHTPINLVRDLPQG
:..* ******:*:.* :*:. *.****:******** **:: :*. : **::***** *
AAP50485.1 FNTLKPIFKLPLGINITNFRAIL----TAFSPAQDI--WGTSAAAYFVGYLKPTTFMLKY
AAP30713.1 FNTLKPIFKLPLGINITNFRAIL----TAFSPAQDI--WGTSAAAYFVGYLKPTTFMLKY
AAP13441.1 FNTLKPIFKLPLGINITNFRAIL----TAFSPAQDI--WGTSAAAYFVGYLKPTTFMLKY
AAP41037.1 FNTLKPIFKLPLGINITNFRAIL----TAFSPAQDI--WGTSAAAYFVGYLKPTTFMLKY
AAU04664.1 FNTLKPIFKLPLGINITNFRAIL----TAFSPAQDT--WGTSAAAYFVGYLKPTTFMLKY
AAV49730.1 FNTLKPIFKLPLGINITNFRAIL----TAFSPAQDT--WGTSAAAYFVGYLKPTTFMLKY
AAV97995.1 FNTLKPIFKLPLGINITNFRAIL----TAFSPAQDT--WGTSAAAYFVGYLKPTTFMLKY
AAV98000.1 FNTLKPIFKLPLGIKITNFRAIL----TAFSPAQGT--WGTSAAAYFVGYLKPTTFMLKY
n6VXX_1|Ch FSALEPLVDLPIGINITRFQTLLALHRSYLTPGDSSSGWTAGAAAYYVGYLQPRTFLLKY
QHR63300.2 FSALEPLVDLPIGINITRFQTLLALHRSYLTPGDSSSGWTAGAAAYYVGYLQPRTFLLKY
sp|P0DTC2| FSALEPLVDLPIGINITRFQTLLALHRSYLTPGDSSSGWTAGAAAYYVGYLQPRTFLLKY
QHD43416.1 FSALEPLVDLPIGINITRFQTLLALHRSYLTPGDSSSGWTAGAAAYYVGYLQPRTFLLKY
*.:*:*:..**:**:**.*.::* : ::*.:. * :.****:****:* **:***
AAP50485.1 DENGTITDAVDCSQNPLAELKCSVKSFEIDKGIYQTSNFRVVPSGDVVRFPNITNLCPFG
AAP30713.1 DENGTITDAVDCSQNPLAELKCSVKSFEIDKGIYQTSNFRVVPSGDVVRFPNITNLCPFG
AAP13441.1 DENGTITDAVDCSQNPLAELKCSVKSFEIDKGIYQTSNFRVVPSGDVVRFPNITNLCPFG
AAP41037.1 DENGTITDAVDCSQNPLAELKCSVKSFEIDKGIYQTSNFRVVPSGDVVRFPNITNLCPFG
AAU04664.1 DENGTITDAVDCSQNPLAELKCSVKSFEIDKGIYQTSNFRVVPSGDVVRFPNITNLCPFG
AAV49730.1 DENGTITDAVDCSQNPLAELKCSVKSFEIDKGIYQTSNFRVVPSGDVVRFPNITNLCPFG
AAV97995.1 DENGTITDAVDCSQNPLAELKCSVKSFEIDKGIYQTSNFRVVPSGDVVRFPNITNLCPFG
AAV98000.1 DENGTITDAVDCSQNPLAELKCSVKSFEIDKGIYQTSNFRVVPSGDVVRFPNITNLCPFG
n6VXX_1|Ch NENGTITDAVDCALDPLSETKCTLKSFTVEKGIYQTSNFRVQPTESIVRFPNITNLCPFG
QHR63300.2 NENGTITDAVDCALDPLSETKCTLKSFTVEKGIYQTSNFRVQPTDSIVRFPNITNLCPFG
sp|P0DTC2| NENGTITDAVDCALDPLSETKCTLKSFTVEKGIYQTSNFRVQPTESIVRFPNITNLCPFG
QHD43416.1 NENGTITDAVDCALDPLSETKCTLKSFTVEKGIYQTSNFRVQPTESIVRFPNITNLCPFG
:***********: :**:* **::*** ::*********** *: .:*************
AAP50485.1 EVFNATKFPSVYAWERKKISNCVADYSVLYNSTFFSTFKCYGVSATKLNDLCFSNVYADS
AAP30713.1 EVFNATKFPSVYAWERKKISNCVADYSVLYNSTFFSTFKCYGVSATKLNDLCFSNVYADS
AAP13441.1 EVFNATKFPSVYAWERKKISNCVADYSVLYNSTFFSTFKCYGVSATKLNDLCFSNVYADS
AAP41037.1 EVFNATKFPSVYAWERKKISNCVADYSVLYNSTFFSTFKCYGVSATKLNDLCFSNVYADS
AAU04664.1 EVFNATKFPSVYAWERKRISNCVADYSVLYNSTSFSTFKCYGVSATKLNDLCFSNVYADS
AAV49730.1 EVFNATKFPSVYAWERKRISNCVADYSVLYNSTSFSTFKCYGVSATKLNDLCFSNVYADS
AAV97995.1 EVFNATKFPSVYAWERKRISNCVADYSVLYNSTSFSTFKCYGVSATKLNDLCFSNVYADS
AAV98000.1 EVFNATKFPSVYAWERKRISNCVADYSVLYNSTSFSTFKCYGVSATKLNDLCFSNVYADS
n6VXX_1|Ch EVFNATRFASVYAWNRKRISNCVADYSVLYNSASFSTFKCYGVSPTKLNDLCFTNVYADS
QHR63300.2 EVFNATTFASVYAWNRKRISNCVADYSVLYNSTSFSTFKCYGVSPTKLNDLCFTNVYADS
sp|P0DTC2| EVFNATRFASVYAWNRKRISNCVADYSVLYNSASFSTFKCYGVSPTKLNDLCFTNVYADS
QHD43416.1 EVFNATRFASVYAWNRKRISNCVADYSVLYNSASFSTFKCYGVSPTKLNDLCFTNVYADS
****** *.*****:**.**************: **********.********:******
AAP50485.1 FVVKGDDVRQIAPGQTGVIADYNYKLPDDFMGCVLAWNTRNIDATSTGNYNYKYRYLRHG
AAP30713.1 FVVKGDDVRQIAPGQTGVIADYNYKLPDDFMGCVLAWNTRNIDATSTGNYNYKYRYLRHG
AAP13441.1 FVVKGDDVRQIAPGQTGVIADYNYKLPDDFMGCVLAWNTRNIDATSTGNYNYKYRYLRHG
AAP41037.1 FVVKGDDVRQIAPGQTGVIADYNYKLPDDFMGCVLAWNTRNIDATSTGNYNYKYRYLRHG
AAU04664.1 FVVKGDDVRQIAPGQTGVIADYNYKLPDDFMGCVLAWNTRNIDATSTGNYNYKYRYLRHG
AAV49730.1 FVVKGDDVRQIAPGQTGVIADYNYKLPDDFMGCVLAWNTRNIDATSTGNYNYKYRYLRHG
AAV97995.1 FVVKGDDVRQIAPGQTGVIADYNYKLPDDFMGCVLAWNTRNIDATSTGNYNYKFRYLRHG
AAV98000.1 FVVKGDDVRQIAPGQTGVIADYNYKLPDDFMGCVLAWNTRNIDATSTGNYNYKYRYLRHG
n6VXX_1|Ch FVIRGDEVRQIAPGQTGKIADYNYKLPDDFTGCVIAWNSNNLDSKVGGNYNYLYRLFRKS
QHR63300.2 FVITGDEVRQIAPGQTGKIADYNYKLPDDFTGCVIAWNSKHIDAKEGGNFNYLYRLFRKA
sp|P0DTC2| FVIRGDEVRQIAPGQTGKIADYNYKLPDDFTGCVIAWNSNNLDSKVGGNYNYLYRLFRKS
QHD43416.1 FVIRGDEVRQIAPGQTGKIADYNYKLPDDFTGCVIAWNSNNLDSKVGGNYNYLYRLFRKS
**: **:********** ************ ***:***:.::*:. **:** :* :*:.
AAP50485.1 KLRPFERDISNVPFSPDGKPCT-PPALNCYWPLNDYGFYTTTGIGYQPYRVVVLSFELLN
AAP30713.1 KLRPFERDISNVPFSPDGKPCT-PPALNCYWPLNDYGFYTTTGIGYQPYRVVVLSFELLN
AAP13441.1 KLRPFERDISNVPFSPDGKPCT-PPALNCYWPLNDYGFYTTTGIGYQPYRVVVLSFELLN
AAP41037.1 KLRPFERDISNVPFSPDGKPCT-PPALNCYWPLNDYGFYTTTGIGYQPYRVVVLSFELLN
AAU04664.1 KLRPFERDISNVPFSPDGKPCT-PPAPNCYWPLRGYGFYTTSGIGYQPYRVVVLSFELLN
AAV49730.1 KLRPFERDISNVPFSPDGKPCT-PPAPNCYWPLRGYGFYTTSGIGYQPYRVVVLSFELLN
AAV97995.1 KLRPFERDISNVPFSPDGKPCT-PPAPNCYWPLRGYGFYTTSGIGYQPYRVVVLSFELLN
AAV98000.1 KLRPFERDISNVPFSPDGKPCT-PPAPNCYWPLRGYGFYTTSGIGYQPYRVVVLSFELLN
n6VXX_1|Ch NLKPFERDISTEIYQAGSTPCNGVEGFNCYFPLQSYGFQPTNGVGYQPYRVVVLSFELLH
QHR63300.2 NLKPFERDISTEIYQAGSKPCNGQTGLNCYYPLYRYGFYPTDGVGHQPYRVVVLSFELLN
sp|P0DTC2| NLKPFERDISTEIYQAGSTPCNGVEGFNCYFPLQSYGFQPTNGVGYQPYRVVVLSFELLH
QHD43416.1 NLKPFERDISTEIYQAGSTPCNGVEGFNCYFPLQSYGFQPTNGVGYQPYRVVVLSFELLH
:*.*******. :.....**. . ***:** *** .* *:*:*************:
AAP50485.1 APATVCGPKLSTDLIKNQCVNFNFNGLTGTGVLTPSSKRFQPFQQFGRDVSDFTDSVRDP
AAP30713.1 APATVCGPKLSTDLIKNQCVNFNFNGLTGTGVLTPSSKRFQPFQQFGRDVSDFTDSVRDP
AAP13441.1 APATVCGPKLSTDLIKNQCVNFNFNGLTGTGVLTPSSKRFQPFQQFGRDVSDFTDSVRDP
AAP41037.1 APATVCGPKLSTDLIKNQCVNFNFNGLTGTGVLTPSSKRFQPFQQFGRDVSDFTDSVRDP
AAU04664.1 APATVCGPKLSTDLIKNQCVNFNFNGLTGTGVLTPSSKRFQPFQQFGRDVSDFTDSVRDP
AAV49730.1 APATVCGPKLSTDLIKNQCVNFNFNGLTGTGVLTPSSKRFQPFQQFGRDVSDFTDSVRDP
AAV97995.1 APATVCGPKLSTDLIKNQCVNFNFNGLTGTGVLTPSSKRFQPFQQFGRDVSDFTDSVRDP
AAV98000.1 APATVCGPKLSTDLIKNQCVNFNFNGLTGTGVLTPSSKRFQPFQQFGRDVSDFTDSVRDP
n6VXX_1|Ch APATVCGPKKSTNLVKNKCVNFNFNGLTGTGVLTESNKKFLPFQQFGRDIADTTDAVRDP
QHR63300.2 APATVCGPKKSTNLVKNKCVNFNFNGLTGTGVLTESNKKFLPFQQFGRDIADTTDAVRDP
sp|P0DTC2| APATVCGPKKSTNLVKNKCVNFNFNGLTGTGVLTESNKKFLPFQQFGRDIADTTDAVRDP
QHD43416.1 APATVCGPKKSTNLVKNKCVNFNFNGLTGTGVLTESNKKFLPFQQFGRDIADTTDAVRDP
********* **:*:**:**************** *.*.* ********::* **:****
AAP50485.1 KTSEILDISPCSFGGVSVITPGTNASSEVAVLYQDVNCTDVSTAIHADQLTPAWRIYSTG
AAP30713.1 KTSEILDISPCSFGGVSVITPGTNASSEVAVLYQDVNCTDVSTAIHADQLTPAWRIYSTG
AAP13441.1 KTSEILDISPCSFGGVSVITPGTNASSEVAVLYQDVNCTDVSTAIHADQLTPAWRIYSTG
AAP41037.1 KTSEILDISPCAFGGVSVITPGTNASSEVAVLYQDVNCTDVSTAIHADQLTPAWRIYSTG
AAU04664.1 KTSEILDISPCSFGGVSVITPGTNASSEVAVLYQDVNCTDVSTLIHAEQLTPAWRIYSTG
AAV49730.1 KTSEILDISPCSFGGVSVITPGTNASSEVAVLYQDVNCTDVSTLIHAEQLTPAWRIYSTG
AAV97995.1 KTSEILDISPCSFGGVSVITPGTNASSEVAVLYQDVNCTDVSTLIHAEQLTPAWRIYSTG
AAV98000.1 KTSEILDISPCSFGGVSVITPGTNASSEVAVLYQDVNCTDVSTLIHAEQLTPAWRIYSTG
n6VXX_1|Ch QTLEILDITPCSFGGVSVITPGTNTSNQVAVLYQDVNCTEVPVAIHADQLTPTWRVYSTG
QHR63300.2 QTLEILDITPCSFGGVSVITPGTNASNQVAVLYQDVNCTEVPVAIHADQLTPTWRVYSTG
sp|P0DTC2| QTLEILDITPCSFGGVSVITPGTNTSNQVAVLYQDVNCTEVPVAIHADQLTPTWRVYSTG
QHD43416.1 QTLEILDITPCSFGGVSVITPGTNTSNQVAVLYQDVNCTEVPVAIHADQLTPTWRVYSTG
:* *****:**:************:*.:***********:*.. ***:****:**:****
AAP50485.1 NNVFQTQAGCLIGAEHVDTSYECDIPIGAGICASYHTVSLL----RSTSQKSIVAYTMSL
AAP30713.1 NNVFQTQAGCLIGAEHVDTSYECDIPIGAGICASYHTVSLL----RSTSQKSIVAYTMSL
AAP13441.1 NNVFQTQAGCLIGAEHVDTSYECDIPIGAGICASYHTVSLL----RSTSQKSIVAYTMSL
AAP41037.1 NNVFQTQAGCLIGAEHVDTSYECDIPIGAGICASYHTVSLL----RSTSQKSIVAYTMSL
AAU04664.1 NNVFQTQAGCLIGAEHVDTSYECDIPIGAGICASYHTVSSL----RSTSQKSIVAYTMSL
AAV49730.1 NNVFQTQAGCLIGAEHVDTSYECDIPIGAGICASYHTVSSL----RSTSQKSIVAYTMSL
AAV97995.1 NNVFQTQAGCLIGAEHVDTSYECDIPIGAGICASYHTVSSL----RSTSQKSIVAYTMSL
AAV98000.1 NNVFQTQAGCLIGAEHVDSSYECDIPIGAGICASYHTVSSL----RSTSQKSIVAYTMSL
n6VXX_1|Ch SNVFQTRAGCLIGAEHVNNSYECDIPIGAGICASYQTQTNSPSGAGSVASQSIIAYTMSL
QHR63300.2 SNVFQTRAGCLIGAEHVNNSYECDIPIGAGICASYQTQTNS----RSVASQSIIAYTMSL
sp|P0DTC2| SNVFQTRAGCLIGAEHVNNSYECDIPIGAGICASYQTQTNSPRRARSVASQSIIAYTMSL
QHD43416.1 SNVFQTRAGCLIGAEHVNNSYECDIPIGAGICASYQTQTNSPRRARSVASQSIIAYTMSL
.*****.**********:.****************:* : *.:.:**:******
AAP50485.1 GADSSIAYSNNTIAIPTNFSISITTEVMPVSMAKTSVDCNMYICGDSTECANLLLQYGSF
AAP30713.1 GADSSIAYSNNTIAIPTNFSISITTEVMPVSMAKTSVDCNMYICGDSTECANLLLQYGSF
AAP13441.1 GADSSIAYSNNTIAIPTNFSISITTEVMPVSMAKTSVDCNMYICGDSTECANLLLQYGSF
AAP41037.1 GADSSIAYSNNTIAIPTNFSISITTEVMPVSMAKTSVDCNMYICGDSTECANLLLQYGSF
AAU04664.1 GADSSIAYSNNTIAIPTNFSISITTEVMPVSMAKTSVDCNMYICGDSTECANLLLQYGSF
AAV49730.1 GADSSIAYSNNTIAIPTNFSISITTEVMPVSMAKTSVDCNMYICGDSTECANLLLQYGSF
AAV97995.1 GADSSIAYSNNTIAIPTNFSISITTEVMPVSMAKTSVDCNMYICGDSTECANLLLQYGSF
AAV98000.1 GADSSIAYSNNTIAIPTNFSISITTEVMPVSMAKTSVDCNMYICGDSTECANLLLQYGSF
n6VXX_1|Ch GAENSVAYSNNSIAIPTNFTISVTTEILPVSMTKTSVDCTMYICGDSTECSNLLLQYGSF
QHR63300.2 GAENSVAYSNNSIAIPTNFTISVTTEILPVSMTKTSVDCTMYICGDSTECSNLLLQYGSF
sp|P0DTC2| GAENSVAYSNNSIAIPTNFTISVTTEILPVSMTKTSVDCTMYICGDSTECSNLLLQYGSF
QHD43416.1 GAENSVAYSNNSIAIPTNFTISVTTEILPVSMTKTSVDCTMYICGDSTECSNLLLQYGSF
**:.*:*****:*******:**:***::****:******.**********:*********
AAP50485.1 CTQLNRALSGIAAEQDRNTREVFAQVKQMYKTPTLKYFGGFNFSQILPDPLKPTKRSFIE
AAP30713.1 CTQLNRALSGIAAEQDRNTREVFAQVKQMYKTPTLKYFGGFNFSQILPDPLKPTKRSFIE
AAP13441.1 CTQLNRALSGIAAEQDRNTREVFAQVKQMYKTPTLKYFGGFNFSQILPDPLKPTKRSFIE
AAP41037.1 CTQLNRALSGIAAEQDRNTREVFAQVKQMYKTPTLKYFGGFNFSQILPDPLKPTKRSFIE
AAU04664.1 CRQLNRALSGIAAEQDRNTREVFVQVKQMYKTPTLKDFGGFNFSQILPDPLKPTKRSFIE
AAV49730.1 CRQLNRALSGIAAEQDRNTREVFVQVKQMYKTPTLKDFGGFNFSQILPDPLKPTKRSFIE
AAV97995.1 CRQLNRALSGIAAEQDRNTREVFVQVKQMYKTPTLKDFGGFNFSQILPDPLKPTKRSFIE
AAV98000.1 CRQLNRALSGIAAEQDRNTREVFVQVKQMYKTPTLKDFGGFNFSQILPDPLKPTKRSFIE
n6VXX_1|Ch CTQLNRALTGIAVEQDKNTQEVFAQVKQIYKTPPIKDFGGFNFSQILPDPSKPSKRSFIE
QHR63300.2 CTQLNRALTGIAVEQDKNTQEVFAQVKQIYKTPPIKDFGGFNFSQILPDPSKPSKRSFIE
sp|P0DTC2| CTQLNRALTGIAVEQDKNTQEVFAQVKQIYKTPPIKDFGGFNFSQILPDPSKPSKRSFIE
QHD43416.1 CTQLNRALTGIAVEQDKNTQEVFAQVKQIYKTPPIKDFGGFNFSQILPDPSKPSKRSFIE
* ******:***.***.**.***.****:****.:* ************* **:******
AAP50485.1 DLLFNKVTLADAGFMKQYGECLGDINARDLICAQKFNGLTVLPPLLTDDMIAAYTAALVS
AAP30713.1 DLLFNKVTLADAGFMKQYGECLGDINARDLICAQKFNGLTVLPPLLTDDMIAAYTAALVS
AAP13441.1 DLLFNKVTLADAGFMKQYGECLGDINARDLICAQKFNGLTVLPPLLTDDMIAAYTAALVS
AAP41037.1 DLLFNKVTLADAGFMKQYGECLGDINARDLICAQKFNGLTVLPPLLTDDMIAAYTAALVS
AAU04664.1 DLLFNKVTLADAGFMKQYGECLGDINARDLICAQKFNGLTVLPPLLTDDMIAAYTAALVS
AAV49730.1 DLLFNKVTLADAGFMKQYGECLGDINARDLICAQKFNGLTVLPPLLTDDMIAAYTAALVS
AAV97995.1 DLLFNKVTLADAGFMKQYGECLGDINARDLICAQKFNGLTVLPPLLTDDMIAAYTAALVS
AAV98000.1 DLLFNKVTLADAGFMKQYGECLGDINARDLICAQKFNGLTVLPPLLTDDMIAAYTAALVS
n6VXX_1|Ch DLLFNKVTLADAGFIKQYGDCLGDIAARDLICAQKFNGLTVLPPLLTDEMIAQYTSALLA
QHR63300.2 DLLFNKVTLADAGFIKQYGDCLGDIAARDLICAQKFNGLTVLPPLLTDEMIAQYTSALLA
sp|P0DTC2| DLLFNKVTLADAGFIKQYGDCLGDIAARDLICAQKFNGLTVLPPLLTDEMIAQYTSALLA
QHD43416.1 DLLFNKVTLADAGFIKQYGDCLGDIAARDLICAQKFNGLTVLPPLLTDEMIAQYTSALLA
**************:****:***** **********************:*** **:**::
AAP50485.1 GTATAGWTFGAGAALQIPFAMQMAYRFNGIGVTQNVLYENQKQIANQFNKAISQIQESLT
AAP30713.1 GTATAGWTFGAGAALQIPFAMQMAYRFNGIGVTQNVLYENQKQIANQFNKAISQIQESLT
AAP13441.1 GTATAGWTFGAGAALQIPFAMQMAYRFNGIGVTQNVLYENQKQIANQFNKAISQIQESLT
AAP41037.1 GTATAGWTFGAGAALQIPFAMQMAYRFNGIGVTQNVLYENQKQIANQFNKAISQIQESLT
AAU04664.1 GTATAGWTFGAGAALQIPFAMQMAYRFNGIGVTQNVLYENQKQIANQFNKAISQIQESLT
AAV49730.1 GTATAGWTFGAGAALQIPFAMQMAYRFNGIGVTQNVLYENQKQIANQFNKAISQIQESLT
AAV97995.1 GTATAGWTFGAGAALQIPFAMQMAYRFNGIGVTQNVLYENQKQIANQFNKAISQIQESLT
AAV98000.1 GTATAGWTFGAGAALQIPFAMQMAYRFNGIGVTQNVLYENQKQIANQFNKAISQIQESLT
n6VXX_1|Ch GTITSGWTFGAGAALQIPFAMQMAYRFNGIGVTQNVLYENQKLIANQFNSAIGKIQDSLS
QHR63300.2 GTITSGWTFGAGAALQIPFAMQMAYRFNGIGVTQNVLYENQKLIANQFNSAIGKIQDSLS
sp|P0DTC2| GTITSGWTFGAGAALQIPFAMQMAYRFNGIGVTQNVLYENQKLIANQFNSAIGKIQDSLS
QHD43416.1 GTITSGWTFGAGAALQIPFAMQMAYRFNGIGVTQNVLYENQKLIANQFNSAIGKIQDSLS
** *:************************************* ******.**.:**:**:
AAP50485.1 TTSTALGKLQDVVNQNAQALNTLVKQLSSNFGAISSVLNDILSRLDKVEAEVQIDRLITG
AAP30713.1 TTSTALGKLQDVVNQNAQALNTLVKQLSSNFGAISSVLNDILSRLDKVEAEVQIDRLITG
AAP13441.1 TTSTALGKLQDVVNQNAQALNTLVKQLSSNFGAISSVLNDILSRLDKVEAEVQIDRLITG
AAP41037.1 TTSTALGKLQDVVNQNAQALNTLVKQLSSNFGAISSVLNDILSRLDKVEAEVQIDRLITG
AAU04664.1 TTSTALGKLQDVVNQNAQALNTLVKQLSSNFGAISSVLNDILSRLDKVEAEVQIDRLITG
AAV49730.1 TTSTALGKLQDVVNQNAQALNTLVKQLSSNFGAISSVLNDILSRLDKVEAEVQIDRLITG
AAV97995.1 TTSTALGKLQDVVNQNAQALNTLVKQLSSNFGAISSVLNDILSRLDKVEAEVQIDRLITG
AAV98000.1 TTSTALGKLQDVVNQNAQALNTLVKQLSSNFGAISSVLNDILSRLDKVEAEVQIDRLITG
n6VXX_1|Ch STASALGKLQDVVNQNAQALNTLVKQLSSNFGAISSVLNDILSRLDPPEAEVQIDRLITG
QHR63300.2 STASALGKLQDVVNQNAQALNTLVKQLSSNFGAISSVLNDILSRLDKVEAEVQIDRLITG
sp|P0DTC2| STASALGKLQDVVNQNAQALNTLVKQLSSNFGAISSVLNDILSRLDKVEAEVQIDRLITG
QHD43416.1 STASALGKLQDVVNQNAQALNTLVKQLSSNFGAISSVLNDILSRLDKVEAEVQIDRLITG
:*::****************************************** ************
AAP50485.1 RLQSLQTYVTQQLIRAAEIRASANLAATKMSECVLGQSKRVDFCGKGYHLMSFPQAAPHG
AAP30713.1 RLQSLQTYVTQQLIRAAEIRASANLAATKMSECVLGQSKRVDFCGKGYHLMSFPQAAPHG
AAP13441.1 RLQSLQTYVTQQLIRAAEIRASANLAATKMSECVLGQSKRVDFCGKGYHLMSFPQAAPHG
AAP41037.1 RLQSLQTYVTQQLIRAAEIRASANLAATKMSECVLGQSKRVDFCGKGYHLMSFPQAAPHG
AAU04664.1 RLQSLQTYVTQQLIRAAEIRASANLAATKMSECVLGQSKRVDFCGKGYHLMSFPQAAPHG
AAV49730.1 RLQSLQTYVTQQLIRAAEIRASANLAATKMSECVLGQSKRVDFCGKGYHLMSFPQAAPHG
AAV97995.1 RLQSLQTYVTQQLIRAAEIRASANLAATKMSECVLGQSKRVDFCGKGYHLMSFPQAAPHG
AAV98000.1 RLQSLQTYVTQQLIRAAEIRASANLAATKMSECVLGQSKRVDFCGKGYHLMSFPQAAPHG
n6VXX_1|Ch RLQSLQTYVTQQLIRAAEIRASANLAATKMSECVLGQSKRVDFCGKGYHLMSFPQSAPHG
QHR63300.2 RLQSLQTYVTQQLIRAAEIRASANLAATKMSECVLGQSKRVDFCGKGYHLMSFPQSAPHG
sp|P0DTC2| RLQSLQTYVTQQLIRAAEIRASANLAATKMSECVLGQSKRVDFCGKGYHLMSFPQSAPHG
QHD43416.1 RLQSLQTYVTQQLIRAAEIRASANLAATKMSECVLGQSKRVDFCGKGYHLMSFPQSAPHG
*******************************************************:****
AAP50485.1 VVFLHVTYVPSQERNFTTAPAICHEGKAYFPREGVFVFNGTSWFITQRNFFSPQIITTDN
AAP30713.1 VVFLHVTYVPSQERNFTTAPAICHEGKAYFPREGVFVFNGTSWFITQRNFFSPQIITTDN
AAP13441.1 VVFLHVTYVPSQERNFTTAPAICHEGKAYFPREGVFVFNGTSWFITQRNFFSPQIITTDN
AAP41037.1 VVFLHVTYVPSQERNFTTAPAICHEGKAYFPREGVFVFNGTSWFITQRNFFSPQIITTDN
AAU04664.1 VVFLHVTYVPSQERNFTTAPAICHEGKAYFPREGVFVFNGTSWFITQRNFFSPQIITTDN
AAV49730.1 VVFLHVTYVPSQERNFTTAPAICHEGKAYFPREGVFVFNGTSWFITQRNFFSPQIITTDN
AAV97995.1 VVFLHVTYVPSQERNFTTAPAICHEGKAYFPREGVFVFNGTSWFITQRNFFSPQIITTDN
AAV98000.1 VVFLHVTYVPSQERNFTTAPAICHEGKAYFPREGVFVFNGTSWFITQRNFFSPQIITTDN
n6VXX_1|Ch VVFLHVTYVPAQEKNFTTAPAICHDGKAHFPREGVFVSNGTHWFVTQRNFYEPQIITTDN
QHR63300.2 VVFLHVTYVPAQEKNFTTAPAICHDGKAHFPREGVFVSNGTHWFVTQRNFYEPQIITTDN
sp|P0DTC2| VVFLHVTYVPAQEKNFTTAPAICHDGKAHFPREGVFVSNGTHWFVTQRNFYEPQIITTDN
QHD43416.1 VVFLHVTYVPAQEKNFTTAPAICHDGKAHFPREGVFVSNGTHWFVTQRNFYEPQIITTDN
**********:**.**********:***:******** *** **:*****:.********
AAP50485.1 TFVSGNCDVVIGIINNTVYDPLQPELDSFKEELDKYFKNHTSPDVDFGDISGINASVVNI
AAP30713.1 TFVSGNCDVVIGIINNTVYDPLQPELDSFKEELDKYFKNHTSPDVDLGDISGINASVVNI
AAP13441.1 TFVSGNCDVVIGIINNTVYDPLQPELDSFKEELDKYFKNHTSPDVDLGDISGINASVVNI
AAP41037.1 TFVSGNCDVVIGIINNTVYDPLQPELDSFKEELDKYFKNHTSPDVDLGDISGINASVVNI
AAU04664.1 TFVSGNCDVVIGIINNTVYDPLQPELDSFKEELDKYFKNHTSPDVDLGDISGINASVVNI
AAV49730.1 TFVSGNCDVVIGIINNTVYDPLQPELDSFKEELDKYFKNHTSPDVDLGDISGINASVVNI
AAV97995.1 TFVSGNCDVVIGIINNTVYDPLQPELDSFKEELDKYFKNHTSPDVDLGDISGINASVVNI
AAV98000.1 TFVSGNCDVVIGIINNTVYDPLQPELDSFKEELDKYFKNHTSPDVDLGDISGINASVVNI
n6VXX_1|Ch TFVSGNCDVVIGIVNNTVYDPLQPELDSFKEELDKYFKNHTSPDVDLGDISGINASVVNI
QHR63300.2 TFVSGSCDVVIGIVNNTVYDPLQPELDSFKEELDKYFKNHTSPDVDLGDISGINASVVNI
sp|P0DTC2| TFVSGNCDVVIGIVNNTVYDPLQPELDSFKEELDKYFKNHTSPDVDLGDISGINASVVNI
QHD43416.1 TFVSGNCDVVIGIVNNTVYDPLQPELDSFKEELDKYFKNHTSPDVDLGDISGINASVVNI
*****.*******:********************************:*************
AAP50485.1 QKEIDRLNEVAKNLNESLIDLQELGKYEQYIKWPWYVWLGFIAGLIAIVMVTILLCCMTS
AAP30713.1 QKEIDRLNEVAKNLNESLIDLQELGKYEQYIKWPWYVWLGFIAGLIAIVMVTILLCCMTS
AAP13441.1 QKEIDRLNEVAKNLNESLIDLQELGKYEQYIKWPWYVWLGFIAGLIAIVMVTILLCCMTS
AAP41037.1 QKEIDRLNEVAKNLNESLIDLQELGKYEQYIKWPWYVWLGFIAGLIAIVMVTILLCCMTS
AAU04664.1 QEEIDRLNEVAKNLNESLIDLQELGKYEQYIKWPWYVWLGFIAGLIAIVMVTILLCCMTS
AAV49730.1 QEEIDRLNEVAKNLNESLIDLQELGKYEQYIKWPWYVWLGFIAGLIAIVMVTILLCCMTS
AAV97995.1 QEEIDRLNEVAKNLNESLIDLQELGKYEQYIKWPWYVWLGFIAGLIAIVMVTILLCCMTS
AAV98000.1 QEEIDRLNEVAKNLNESLIDLQELGKYEQYIKWPWYVWLGFIAGLIAIVMVTILLCCMTS
n6VXX_1|Ch QKEIDRLNEVAKNLNESLIDLQELGKYEQYIKGSGRENLYFQGG---------------G
QHR63300.2 QKEIDRLNEVAKNLNESLIDLQELGKYEQYIKWPWYIWLGFIAGLIAIIMVTIMLCCMTS
sp|P0DTC2| QKEIDRLNEVAKNLNESLIDLQELGKYEQYIKWPWYIWLGFIAGLIAIVMVTIMLCCMTS
QHD43416.1 QKEIDRLNEVAKNLNESLIDLQELGKYEQYIKWPWYIWLGFIAGLIAIVMVTIMLCCMTS
*:****************************** . .* * .* .
AAP50485.1 CCSCLKGACSCGSCCKFDEDDSEPVLKGVKL------HYT
AAP30713.1 CCSCLKGACSCGSCCKFDEDDSEPVLKGVKL------HYT
AAP13441.1 CCSCLKGACSCGSCCKFDEDDSEPVLKGVKL------HYT
AAP41037.1 CCSCLKGACSCGSCCKFDEDDSEPVLKGVKL------HYT
AAU04664.1 CCSCLKGACSCGSCCKFDEDDSEPVLKGVKL------HYT
AAV49730.1 CCSCLKGACSCGSCCKFDEDDSEPVLKGVKL------HYT
AAV97995.1 CCSCLKGACSCGSCCKFDEDDSEPVLKGVKL------HYT
AAV98000.1 CCSCLKGACSCGSCCKFDEDDSEPVLKGVKL------HYT
n6VXX_1|Ch GSGYIPEAPRDGQA--YVRKDGEWVLLSTFLGHHHHHHHH
QHR63300.2 CCSCLKGCCSCGSCCKFDEDDSEPVLKGVKL------HYT
sp|P0DTC2| CCSCLKGCCSCGSCCKFDEDDSEPVLKGVKL------HYT
QHD43416.1 CCSCLKGCCSCGSCCKFDEDDSEPVLKGVKL------HYT
.. : . *.. : .*.* ** .. * *:
- Symbols denoting differences between sequences:
- * = perfect alignment
- : = strong similarity
- . = weak similarity
- - no similarity
- Multiple regions showed non-perfect alignments
- Key regions characterized by the amount of non "*" symbols
- Receptor binding domain. RBD of spike protein was the area of interest
- RBD is what interacts with ACE2
- RBD is bold in sequence alignment above
- Chosen sequence with highlighted regions of most difference:

- SARS-CoV epidemic and reemergence – identical in the highlighted regions
- Between SARS-CoV and SARS-CoV-2 – 9/13 are changed
- S to Q – Serine(Polar) to Glutamine(Polar)
- P to A – Proline(Polar) to Alanine(nonpolar)
- D to G – Aspartic Acid(negative) to Glycine(nonpolar)
- G to S – Glycine(nonpolar) to Serine(Polar)
- K to T – Lysine(positive) to Threonine(Polar)
- Y to Q – Tyrosine(nonpolar) to Glutamine(Polar)
- T to P – Threonine(Polar) to Proline(Polar)
- S to N – Serine(Polar) to Asparagine(Polar)
- I to V – Isoleucine(nonpolar) to Valine(nonpolar)
- More changes from nonpolar to polar in beta-3-4 sheet
- SARS-CoV epidemic and reemergence are identical
- Suggests that strength of RBD-ACE2 interaction does not influence pathogenicity
Phylogenetic Tree
- Phylogenetic tree created using the chosen S protein sequences
- Tree was generated on www.phylogeny.fr
- Used "One Click" under "Phylogeny Analysis"
- Uploaded FASTA files of all sequences and pressed "submit"
- Phylogenetic tree for the sequences chosen for this study:
- Tree shows the relationship between the three SARS viruses
- Sequences for each virus cluster together
- The SARS strains from 2002 and 2003-04 form their own clade before splitting of into a clade for each virus
- Suggests that the SARS-CoV strains are more closely related to each other than they are to strains from SARS-CoV-2
- Tree suggests notable sequence differences among different strains
- Bat sequence(QHR63300.2) in its own clade
- Likely due to virus originating in bats
- Bat sequence(QHR63300.2) in its own clade
Visualization of Protein Structure
- Virus IDs searched in NCBI, structures visualized in NCBI Structure viewer iCn3D
- 6VXX from Walls et al.(2020) shows regions in S Protein with high density of differences
- Full S protein with areas of high amount of of differences marked
- RBD region boxed in red and highlighted in yellow
- Other notable regions circled in red
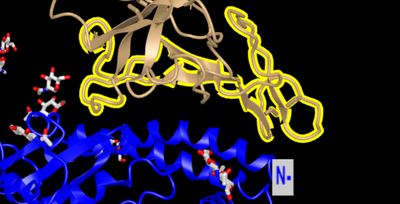
- 6M17 from Yan et al.(2020) shows RBD-ACE2 interaction
- Regions from RBD that had a large amount of differences were were highlighted
- Shows region in RBD with large amount of differences
- RBD is brown and ACE2 is blue
- Region of interest is highlighted in yellow
- Region interacts with alpha1, alpha2, and beta3-4 loop(Yan et al., 2020)
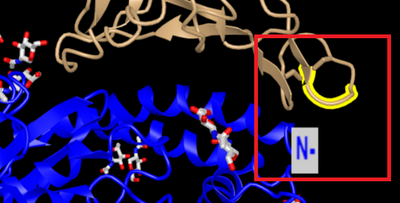
- Shows region in sequence from RBD with a very high difference in amino acids
- Region of interest is highlighted in yellow and boxed in red
- Area that is highlighted interacts with alpha1 N-terminus, which is an area of high interaction
- Shows region in sequence from RBD with a very high difference in amino acids
- Region of interest is highlighted in yellow and boxed in red
- Area that is highlighted interacts with beta3-4 loop, which is a secondary interaction site
Data and Files
- SARS-CoV-2 S protein reference sequence
- SARS epidemic(2002-2003) S sequences
- SARS reemergence(2004) S sequences
- SARS-CoV-2 S sequences
- Phylogenetic tree
Presentation
File:Coronavirus Structure Research Project.pdf
Scientific Conclusion
This research project set out to determine if the variations in the amino acids of the spike glycoprotein were responsible for the infectiousness of the virus. It was found that although the mortality of the SARS reemergence was much lower than that of the epidemic virus strain, the sequences were virtually identical, suggesting that differences in interactions in a different region of interest may be the cause of this. Furthermore, the phylogenetic tree and multiple sequence alignment show amino acid differences between SARS-CoV and SARS-CoV-2, which show that the interactions of the virus with key areas on hACE2 are stronger, which translates to the increased transmissibility of the virus.
Acknowledgements
- My homework partners for this week were Jack Menzagopian and Drew Cartmel
- I referenced Jack's page to get several figures
- The protocol found on BIOL368/S20:Week 14 was used
- Some SARS-CoV-2 sequences were taken from BIOL368/S20:Week 13
- Sequences were obtained from NCBI and UniProt
- Accession numbers for SARS-CoV epidemic and reemergence were obtained from Table 1 in Kan et al.
- Except for what is noted above, this individual journal entry was completed by me and not copied from another source
Nyeo2 (talk) 23:53, 29 April 2020 (PDT)
References
- Kan, B., Wang, M., Jing, H., Xu, H., Jiang, X., Yan, M., ... & Cui, B. (2005). Molecular evolution analysis and geographic investigation of severe acute respiratory syndrome coronavirus-like virus in palm civets at an animal market and on farms. Journal of virology, 79(18), 11892-11900.
- OpenWetWare. (2020). BIOL368/S20:Week 13. Retrieved April 28, 2020 from https://openwetware.org/wiki/BIOL368/S20:Week_13.
- OpenWetWare. (2020). BIOL368/S20:Week 14. Retrieved April 28, 2020 from https://openwetware.org/wiki/BIOL368/S20:Week_14.
- Phylogeny.fr. (2020). Retrieved April 28, 2020 from http://www.phylogeny.fr/.
- Walls, A. C., Park, Y. J., Tortorici, M. A., Wall, A., McGuire, A. T., & Veesler, D. (2020). Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. DOI: 10.1016/j.cell.2020.02.058
- NCBI structure (2020). Retrieved April 29, 2020 from https://www.ncbi.nlm.nih.gov/Structure/index.shtml