Nathan R Beshai Week 5
From OpenWetWare
Jump to navigationJump to search
Nathan R. Beshai User Page
Nathan R. Beshai Template Page
Course assignments
Individual journal assignments
- Nathan R Beshai Week 2
- Nathan R Beshai Week 3
- Nathan R Beshai Week 4
- Nathan R Beshai Week 5
- Nathan R Beshai Week 6
- Nathan R Beshai Week 7
- Nathan R Beshai Week 8
- Nathan R Beshai Week 9
- Nathan R Beshai Week 10
- Nathan R Beshai Week 11
- The D614G Research Group Week 12
- The D614G Research Group Week 14
Class Journals
- Class Journal 1
- Class Journal 2
- Class Journal 3
- Class Journal 4
- Class Journal 5
- Class Journal 6
- Class Journal 7
- Class Journal 8
- Class Journal 9
- Class Journal 10
- Class Journal 11
- Class Journal 12
- Class Journal 14
Link to Brightspace and LMU's Homepage
Purpose
- Exploring the ACE-2 and Spike protein structures, interactions, and classifications through viewing 3-dimensional protein shapes through the NCBI protein database, which helps understand protein binding better.
Methods and Results
Exploring the Spike protein Structure
- Visited the UniProt Knowledgebase (UniProt KB) and searched for the keyword Sars-CoV-2 to answer the following questions.
- Searched using the keyword "SARS-CoV-2" in the main UniProt search field. Answered how many results do you get? Are all the results viral proteins?
- There were 1,616 results that came up for the keyword "SARS-CoV-2". 18 of them were reviewed Swiss-Prot and the rest were unreviewed.
- They were not all viral proteins. A quick glance showed some were Mouse proteins and others were Human proteins such as the human ACE-2 proteins.
- Searched using the keyword "SARS-CoV-2" in the main UniProt search field. Answered how many results do you get? Are all the results viral proteins?
- Searched through the database again and pulled up the spike protein for SARS-CoV-2 through its accession number "P0DTC2".
- Answered: what types of information are provided about this protein in this database entry?
- The type of information that is listed is the:
- Function: UniPro lists the functions of the spike protein s1, spike protein s2, spike protein s3, and the spike glycoprotein.
- The database also lists the molecular function and biological process, its names and taxonomy, its subcellular location, its Pathology, and Biotech, it's PTM/ Processing, How it interacts, its structure, it's family and domains, its sequence, similar proteins, and cross-references.
- The type of information that is listed is the:
- Answered: what types of information are provided about this protein in this database entry?
- Opened the NCBI Structure database using a web browser and chose the SARS-CoV RBD (the year 2002) complexed with Human ACE-2 to work on.
- Used the accession number is 2AJF.
- Pasted the image below.
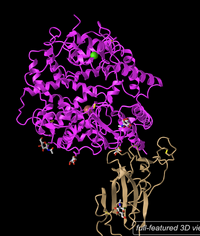
Figure 1: 3d image of SARS-CoV RBD (the year 2002) complexed with Human ACE-2 (2AJF) (NCBI).- Answered the following questions.
- Is this a primary, secondary, tertiary, or quaternary structure? Explain your reasoning.
- The image shows a quaternary structure. This can be seen because there are two peptides that are bound to each other. This can be seen because each peptide has its own N-termini and c-termini indicating a separate peptide.
- How many domains are in this structure? Explain your reasoning.
- There are two domains for this structure because there are two distinct proteins with different functions bound together. This can be seen because the peptides have their own side chains and one of them consists of anti-parallel beta-sheets and the other of alpha-helices. The ACE-2 is the receptor and the Spike RBD which is the binder.
- Is this a primary, secondary, tertiary, or quaternary structure? Explain your reasoning.
- Edited the style and the protein to show the protein as a cylinder and plate style. Copied and pasted the figure below. Answered what is unique to the style.
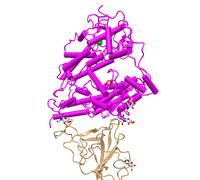
- Figure 2: 3d image of SARS-CoV RBD (the year 2002) complexed with Human ACE-2 (2AJF) edited with the cylinder and plate style(NCBI).
- This style shows the protein the same with the alpha-helices as cylinders and the beta-pleated sheets as plates. This simplified the model and distinguished the secondary structures as shapes instead of stringed peptides.
- Edited the style and the protein to show the protein as a C Alpha Trace style. Copied and pasted the figure below. Answered what is unique to the style.
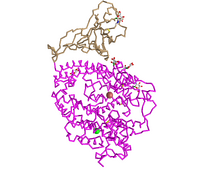
- Figure 3: 3d image of SARS-CoV RBD (the year 2002) complexed with Human ACE-2 (2AJF) edited with the C-alpha trace style.
- This style takes out the secondary structures and just showed connecting alpha carbons connected. This helped put an estimation for the number of alpha carbons in the quaternary structure. This also shows the backbone structure and the rigid shape of the secondary structures.
- Edited the style and the protein to show the protein as a lines style. Copied and pasted the figure below. Answered what is unique to the style.

- Figure 4: 3d image of SARS-CoV RBD (the year 2002) complexed with Human ACE-2 (2AJF) edited with the lines style.
- This syle shows the peptide bonds and side items as a faze in the background and highlighted the 5 amino acid side chains that were bound together. This showed the 5 Human-ACE-2 amino acids with the 5 SARS-COV RBD amino acids bound together.
- Edited the style and the protein to show the protein as a ball and stick. Copied and pasted the figure below. Answered what is unique to the style.

- Figure 5: 3d image of SARS-CoV RBD (the year 2002) complexed with Human ACE-2 (2AJF) edited with the ball and stick style.
- This style helped the viewer see the peptides with their side atoms connecting to the peptide backbone, as well as the amino acid sidechains connected to the backbone.
- Edited the style and the protein to show the protein as a sphere. Copied and pasted the figure below. Answered what is unique to the style.
- Edited the color of the sphere style and the protein to show the protein spectrum colored. Copied and pasted the figure below. Answered what is unique each of the color schemes.
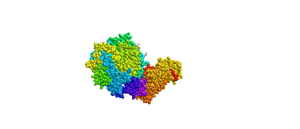
- Figure 7: 3d image of SARS-CoV RBD (the year 2002) complexed with Human ACE-2 (2AJF) edited with the sphere style and spectrum colored.
- The purpose of this model is to show how the type of amino acids is usually together on the peptide. The colors represent different groups of the peptide, however, NCBI does not specify their separate classifications.
- Edited the color of the sphere style and the protein to show the secondary structures colored with beta-sheets green and alpha helices red. Copied and pasted the figure below. Answered what is unique each of the color schemes.
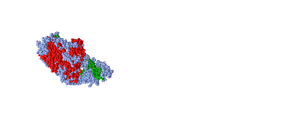
- Figure 8: 3d image of SARS-CoV RBD (the year 2002) complexed with Human ACE-2 (2AJF) edited with the sphere style with the secondary structures highlighted with the beta-sheets green and the alpha-helices in red.
- This model is good to show how each peptide preferred a specific secondary structure. One peptide is all alpha helices and the other is all beta-pleated sheets or random coils.
- Edited the color of the sphere style and the protein to show the protein charges colored. Copied and pasted the figure below. Answered what is unique each of the color schemes.

- Figure 9: 3d image of SARS-CoV RBD (the year 2002) complexed with Human ACE-2 (2AJF) edited with the sphere style and charged amino acids highlighted.
- This style is useful for where the charged amino acids are present. This can give insight to where they are located. Charged amino acids tend to be on the surface, hydrogen bonding with water. This is useful because it shows the hydrophobic amino acids in the center.
- Edited the color of the sphere style and the protein to show the protein atoms colored. Copied and pasted the figure below. Answered what is unique each of the color schemes.

- Figure 10: 3d image of SARS-CoV RBD (the year 2002) complexed with Human ACE-2 (2AJF) edited with the sphere style with the atoms colored. Red signifies an oxygen atom and blue signifies a nitrogen atom.
- This color scheme is useful to show the oxygens present in the atom and nitrogens present in the atom. This helps see the oxygens and nitrogens in the backbone and those in the amino acids. It is also to show the amino acid and oxygen-hydrogen bonding between peptides whether between every four amino acids on the alpha helices or between the antiparallel beta-sheets.
- Took the screenshot of the SARS-CoV 2002 Humam ACE-2 and Spike protein RBD and placed it into a note drawing app and highlighted the N-Terminus and C-Terminus by locating the Carbons on the nearest Beta sheet with the arrow pointed at the alpha carbon. On the other peptide, the c-terminus was located by looking at the last alpha helices and seeing the carbon kink was away from the edge pointing to the n-terminus and making the other end the c-terminus.
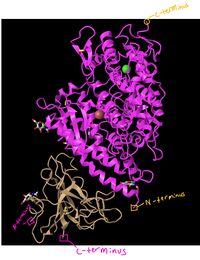
- Figure 11: 3d image of SARS-CoV RBD (the year 2002) complexed with Human ACE-2 (2AJF) with the N-terminus and c-terminus highlighted.
- Answered What secondary structures are present in the ACE-2 Protein and how this was found.
- The secondary structures present in the ACE-2 protein are alpha-helices, random coils, and a couple of rare beta-sheets. This can be seen by a close examination of the ACE-2 structure and the cylinder structures in Figure 2 and the twisted structures in the rest of the figures are alpha-helices. There are random coils between the alpha helices which just give space for the next secondary structure. Lastly, a couple of antiparallel beta sheets can be seen. this can be seen by the arrow symbols which indicate the direction the beta-sheet is pointing.
- Answered What secondary structures are present in the Spike Protein and how this was found.
- The only structures present in the spike protein is antiparallel beta-pleated sheets and random coils between the sheets. This can be seen by the arrows in the figures and green coloring in figure 8 which indicates beta-sheets. The arrows indicate which direction the beta-sheet is pointing. The random coils are present and can be seen as strings between the different structures.
- Looked at side chain-side chain interactions in the civet ACE2-spike protein structure shown in Figure 4B of the Wan et. al. 2020 article.
- Clicked on the link to interact with the structure in iCn3D.
- Clicked on the Windows menu to “View Sequences & Annotations”. In the new window that appears to the right, we clicked on the “Details” tab to show the actual amino acid sequences. There are 2 sets of ACE2-spike proteins because of the way the proteins crystallized. Focused on the pink and tan chains and orient them like is shown in Figure 4B of the article.
- Made the amino acid side chains are shown in the figure visible.
- In the sequence, the window went to sequence “Protein 3SCK_A” (in pink) and select the following amino acids
- T31
- E35
- E38
- T82
- K353
- The part of the ribbon that represents these amino acids should be highlighted in yellow in the structure.
- Went to the styles menu and selected Proteins > Ball and Stick
- Went to the Color menu and selected Atom.
- Saw the side chains shown in the figure.
- Repeated this process for the tan spike protein sequence 3SCK_E for the following amino acids:
- T487
- R479
- G480
- Y442
- P472
- Took screenshot and placed it below.
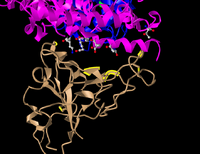
Figure12: 3d structure for the tan spike protein sequences and civet ACE2 with the bound amino acids. - Answered the following questions regarding the figure 12.
- E35 on ACE2 makes an ionic bond with R479 on spike protein. Which of these is acidic and which is basic? How can you tell that from the image you created?
- The E35 amino acid was the basic amino acid and the R479 was the acidic amino acid. This can be seen because the E35 had two oxygens at the end. The oxygen can only be negatively charged and is the hydrogen acceptor. This means that to be hydrogen bond to the R479, that amino acid had to be acidic because it needs a hydrogen doner from the acidic side chain.
- T31 on ACE2 makes a hydrogen bond with Y442 on spike protein. Which side-chain classification do these two amino acids belong to (acidic, basic, uncharged polar, nonpolar)? How can you tell that from the image you created?
- The two hydrogen-bonded amino acids are uncharged polar which can be seen because they are hydrogen-bonded. In order to hydrogen bond, the hydrogen of the Y442 is donated to the carbonyl group of the T31 amino acid. If these were not polar they could not hydrogen bond and if they were charged they could not be hydrogen-bonded.
- Made the dashed lines for these bonds in iCn3D by:
- Going to the View menu and select H bonds & Interactions
- In part 1 of the window that appears, we uncheck “Contacts/Interactions” leaving Hydrogen Bonds and Ionic Interaction checked
- In part 2 of the window, we selected the first set “3SCK_A” (pink)
- In part 3 of the window, we selected the second set “3SCK_E” (tan)
- In part 4 of the window, we click the button “3D Display”
- Saw the dashed lines representing the ionic bonds and H-bonds between the two polypeptide chains and amino acids we described above.
- Took a screenshot and pasted it below.
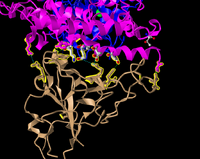
Figure13: 3d structure for the tan spike protein sequences and civet ACE2 with the bound amino acids and added the hydrogen bonds.
Introduction to research project
- Answered the following questions regarding the research project.
- What question will you answer about sequence-->structure-->function relationships in the spike and/or ACE2 protein?
- How do ACE2 proteins across multiple species compare and what common mutations and sequence similarities are there.
- What sequences will you use?
- We will be studying the ACE2 sequences for the following animals:
- Human
- Mouse
- Rat
- Pig
- Chimpanzee
- Civet
- Bat
- Ferrats
- Rabbits
- Cats
- We will be studying the ACE2 sequences for the following animals:
- What question will you answer about sequence-->structure-->function relationships in the spike and/or ACE2 protein?
Scientific conclusion
- In this lab, we explored the 3-dimensional structure of proteins and in specific the ACE2 receptor and Spike protein bonding. Using the NCBI Structure database, we were able to view protein structures, bonds, and chemical elements. Looking at the SARS-CoV RBD(2002)- Human-ACE2 complex, we were able to look at different protein styles that helped make inferences about the protein structure and interactions by highlighting certain aspects of the protein and blurring out the non-focused area. The types of secondary structures present in the proteins are also highlighted in the 3d proteins. 3d models of proteins also helped point out the hydrogen bonding of the civet-ACE2 and amino acid interactions. This helped specify the side group classifications and charges. Accessing and viewing 3d-protein structures help biologists make out protein structures and highlight interactions and classifications.
Acknowledgments
- Referenced and copied OpenWebWare syntax from the BIOL368/F20 week 1 page.
- Referenced and copied questions and methods from the BIOL368/F20 week 5 page.
- Referenced MediaWiki for image formatting syntax.
- Worked with my partner Macie Duran on the research question we are studying and the sequences we will study.
- Worked with Dr. Dahlquist, in class, on our research question and sequences to study.
- Referenced the article "Receptor Recognition by the Novel Coronavirus from Wuhan: an Analysis Based on Decade-Long Structural Studies of SARS Coronavirus" for information regarding 3-dimensional proteins.
- Referenced the NCBI protein database for the structures for the RBD (year 2002) complexed with human ACE2 complex 2AJF and the ACE2-spike protein structure, editing and taking screenshots of the sequences.
- Referenced the protein game Fold it for the class journal.
References
- OpenWetWare. (2020). BIOL368/F20:Week 1. Retrieved September 22, 2020, from https://openwetware.org/wiki/BIOL368/F20:Week_1
- OpenWetWare. (2020). BIOL368/F20:Week 5. Retrieved October 07, 2020, from https://openwetware.org/wiki/BIOL368/F20:Week_5
- Wan, Y., et al. (2020). Receptor Recognition by the Novel Coronavirus from Wuhan: an Analysis Based on Decade-Long Structural Studies of SARS Coronavirus. Journal of Virology, 54 (7), retrieved from https://doi.org/10.1128/JVI.00127-20.
- NCBI Structure (2020). SARS-CoV RBD (year 2002) complexed with human ACE2 complex 2AJF, Retrieved from https://www.ncbi.nlm.nih.gov/Structure/icn3d/full.html?&mmdbid=35213&bu=1&showanno=1.
- NCBI Structure (2020). Civet ACE2-Spike protein structure, Retrieved from https://www.ncbi.nlm.nih.gov/Structure/icn3d/full.html?pdbid=%203SCK.
- Fold it Solving Puzzles for Science (2020). The Science Behind Foldit, retrieved from https://fold.it/portal/info/about#folditpub.
"Except for what is noted above, this individual journal entry was completed by me and not copied from another source" Nathan R. Beshai (talk) 22:39, 7 October 2020 (PDT)
