Mpaniag1 Week 14
Purpose
- The purpose of this week's lab was to use information from Wan et al. (2020) paper and use it to develop the question, Can 2019-nCoV bind to certain species ACE2 receptor? and use sequence analysis and structure-function relationship knowledge to answer that question for 5 different species.
Introduction
- Wan et al. (2020) used the information they obtained about the SARS-CoV virus and made a model based on their predictions about how 2019-nCoV would bind to the ACE2 receptor
- The reason they believe 2019-nCoV uses the ACE2 receptor is because SARS-Cov and 2019-nCoV have 77% sequence similarities
- Wan et al. (2020) found five residues they believe would be important for the binding to occur
- Changes in these certain residues of ACE2 can either limit the binding of 2019-nCoV or enhance it
- From their predictions they found that 2019-nCoV would not be able to bind to mouse and rat ACE2 receptors
Methods and Results
Sequence Collection
- Collect ACE2 receptor sequences from 19 different species using Uniprot [1]
- Search ACE2 in the search bar of UniProt
- Species included mammals, reptiles, birds, fish
- Species from the Wan et al. (2020) paper that were used-Civet, Bat, Mouse, Rat, Pig, Orangutan
- New species that have not been tested in the Wan et al. (2020) paper-Platypus, elephant, sperm whale, pig, horse, giant panda, dog, red fox, chickens, turtles, chameleons, zebrafish, opossum
- Click on the entry number of the species
- Click on the sequence tab
- Download FASTA file
- Repeat for all species
Phylogenic Tree
- Go to Phylogeny.fr [2]
- Click on the tab Phylogeny Analysis
- Click on the One-Click Option
- Click on the tab Phylogeny Analysis
- Copy and Paste all sequences from the sequence collection into the box given
- Click Submit
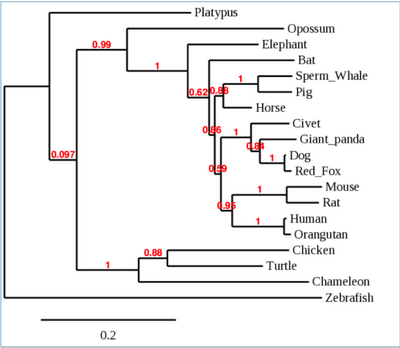
- Take a screenshot of the Tree
- The tree that was made from the species we chose is pictured below
- No patterns were observed from this phylogenetic tree in regard to species that have ACE2 receptors that 2019-nCoV can bind to and those it cannot
Sequence Analysis
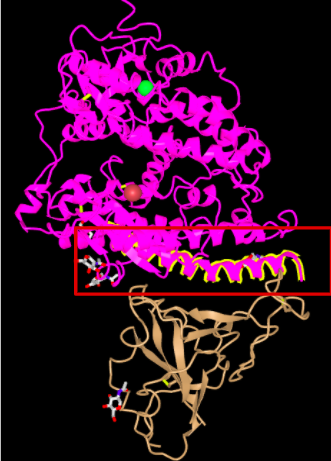
- iCn3D was used to find the sequence of the alpha helix that of ACE2 that 2019n-CoV would bind to
- Go to Protein Data Bank Protein Data Bank and search the PDB ID 2AJF
- Click the sequence tab
- Download the ACE2 sequence
- Go to NCBI Structure home page and search the PDB ID 2AJF
- Enter in the sequence STIEEQAKTFLDKFNHEAEDLFYQSSLASWNYN
- The Alpha Helix closest to the 2019-nCoV will be highlighted
- Photo of the Alpha Helix highlighted below
- Enter in the sequence STIEEQAKTFLDKFNHEAEDLFYQSSLASWNYN
- The sequences of the 19 species were compared using Phylogeny.fr [3]
- Enter in the sequences used for the phylogenetic
- Click Submit
- Click on the tab Alignment
- Click on outputs and choose clustal format
- Using the screenshot tool, snap a picture of the sequence alignment of the 19 species from STIEEQAKTFLDKFNHEAEDLFYQSSLASWNYN
- The sequence for humans ACE2 mentioned above STIEEQAKTFLDKFNHEAEDLFYQSSLASWNYN from the helix was compared to an animal that 2019-nCoV does not bind to, a mouse
- 6 Residue Changes were found that could change the binding, highlighted and Shown Below
- Compare all 17 other sequences to the 2 (mice and humans)
- Find Residues that differ from one species to another
- PDF Below, Yellow (in common with mouse), Green (in common with human), Blue (unique)
File:MAP 19 Sequences Comparison.pdf
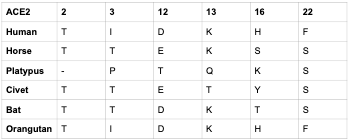
- Choose 5 species to compare with human specifically to see if 2019-nCoV would be able to bind with their ACE2 receptor
- The 5 our research group chose was horse, orangutan, platypus, civet, and bat
Species Sequence Changes in Comparison to Humans
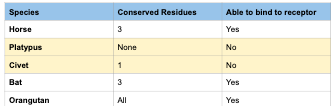
- Horse-4 out of 6 residues were changed
- I->T=non polar to polar
- D->E=both - charged
- H->S=+ charged to polar
- F->S= nonpolar to polar
- Platypus-6 out of 6 residues were changed
- T->-=loss of amino acid
- I->P=both hydrophobic, change of shape
- D->T,= - charged to uncharged
- K->Q= + charged to uncharged
- H->K= both + charged, change of shape
- F->S= hydrophobic to polar
- Civet-5 out of 6 residues were changed
- I->T = nonpolar to polar
- D-> E = both - charged
- K->T = + charge to polar
- H->Y = + charge to polar
- F-> S = nonpolar to polar
- Bat-3 out of 6 residues were changed
- I-> T = nonpolar to polar
- H-> T = + charged to polar
- F-> S = nonpolar to polar
- Orangutan-All residues were conserved
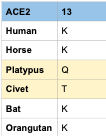
Sequence Analysis with a Focus on position 13
- In Wan et al. (2020) position 31 was considered integral in binding
- In our sequences position 13 correlates with position 31 in Wan et al.'s paper
Comparison of Position 13 amino acids
- If we only compare that residue these are the conclusions that can be made
- Human, Horse, Bat, and Orangutan ACE2 receptors all have the possibility of 2019-nCoV binding with them
- Civet and Platypus ACE2 receptors are less likely to have the possibility of 2019-nCoV binding with them
- If we only compare that residue these are the conclusions that can be made
Results summarized
- Phylogenetic Tree showed no patterns in regard to 2019-nCoV being able to bind to certain ACE2 receptors
- ACE2 receptor's from species to species varied
- If more than three residues of the six were conserved in comparison to humans we made the conclusion that that specified species would be likely to bind to the ACE2 receptor.
Discussion and Future Research
- We wanted to compare protein sequences to see if we could make predictions about the species 2019-nCoV could bind to
- Something to note-we only used the helix directly next to where the 2019-nCoV would bind, but other things could influence that binding
- Several big cats can get 2019-nCoV
- However, civets have been found to have less than ideal binding situations with the 2019-nCoV
- But, civets can get SARS-CoV1
- Future research could include looking why SARS-CoV1 could infect civets but not 2019-nCoV
- However, civets have been found to have less than ideal binding situations with the 2019-nCoV
PowerPoint Presentation
File:MAP Final Presentation Bioinformatics.pdf
Data and Files
- File:Phyl Tree 19 MAP.png
- File:ICn3D structure .png
- File:MAPSequence Analysis.png
- File:MAPResidue Change.png
- File:MAP Position 13.png
- File:MAP 19 Sequences Comparison.pdf
- File:MAP Differences Amino .png
- File:MAP Critera binding.png
- File:MAP Final Presentation Bioinformatics.pdf
Conclusion
In conclusion, during our research, we found that Horse, Bat, and Orangutan all have ACE2 receptors that 2019-nCoV would be likely to bind to. Horse, Bat, and Orangutan all have 3 or more conserved residues when comparing them to six specific residues in humans that were found on the helix closest to where 2019-nCoV would bind. In addition, something they all have in common is at position 13, they all have K while Platypus and Civet have different amino acids and in the Wan et al (2020) paper he stated the amino acid position corresponding to the position 13 is integral for binding. Platypus and Civet are unlikely to have 2019-nCoV bind to their ACE2 receptors.
References
- Home - ORFfinder - NCBI. (n.d.). Retrieved April 22, 2020, from https://www.ncbi.nlm.nih.gov/orffinder/
- NCBI Protein Domains and Macromolecular Structures. (n.d.). Retrieved April 27, 2020, from https://www.ncbi.nlm.nih.gov/Structure/index.shtml
- Phylogeny.fr: Home. (n.d.). Retrieved April 27, 2020, from http://www.phylogeny.fr/
- OpenWetWare. (2020). BIOL368/S20:Week 14. Retrieved April 27 2020, from https://openwetware.org/wiki/BIOL368/S20:Week_14
- RCSB Protein Data Bank. (n.d.). Homepage. Retrieved April 27, 2020, from https://www.rcsb.org/
- UniProtConsortiumEuropean Bioinformatics InstituteProtein Information ResourceSIB Swiss Institute of Bioinformatics. (n.d.). UniProt Consortium. Retrieved April 27, 2020, from http://www.uniprot.org/
- Wan, Y., Shang, J., Graham, R., Baric, R. S., & Li, F. (2020). Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. Journal of virology, 94(7). DOI: 10.1128/JVI.00127-20.
Acknowledgements
- I worked with my homework partners Madeleine B. King, Lizzy Urbina, and Karina Vescio in class to develop the research project, analyze data, and outside of class individually worked on the powerpoint, meeting on Wednesday, April 29, 2020, to practice
- I copied and modified the protocol for Week 14 assignment
- I asked Kam D. Dahlquist, Ph.D. about how to set up the research problem during class on April 23, 2020
- The premise for this research project came from reading Wan et al. 's (2020) paper, Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus.
- Except for what is noted above, this individual journal entry was completed by me and not copied from another source.
Mpaniag1 (talk) 15:01, 29 April 2020 (PDT)
Classwork
Assignments
Journals
- mpaniag1 Week 2
- mpaniag1 Week 3
- mpaniag1 Week 4
- mpaniag1 Week 5
- mpaniag1 Week 6
- mpaniag1 Week 7
- mpaniag1 Week 8
- mpaniag1 Week 9
- mpaniag1 Week 10
- mpaniag1 Week 11
- mpaniag1 Week 13
- mpaniag1 Week 14