Mpaniag1 Week 13
Purpose
The purpose of this week's lab was to discover how to analyze a 3D model of a protein from a sequence, then using the information in different academic journals, make connections about the bonds and orientation to better understand the papers and the differences between them.
Exploring the Spike Protein Structure
The information and models presented below are generated from the amino acid sequence and images found in Wan et al.'s paper "Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus" Wan, Y., Shang, J., Graham, R., Baric, R. S., & Li, F. (2020). Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. Journal of virology, 94(7). DOI: 10.1128/JVI.00127-20
Part 1
- Copy and Paste DNA sequence of spike protein into ExPASY Translate tool
- 6 Different reading frames will appear (pictured below)
- Highlighted in RED will be the open reading frame
- Select the protein sequence that has the most highlighted red sequence
- Highlighted in RED will be the open reading frame
- The correct sequence is 5'-3' Frame 1, I know this because the entire sequence is highlighted in red which means it has the ability to be translated
- Check your answer with the NCBI protein record
- Update answer is correct
Part 2
- Search UniProt Knowledgebase (UniProt KB) and insert the spike protein's DNA sequence provided on the Week 13 Page [1]
- When searching "SARS-CoV" into the UniPort Database 818 results show up as of April 21, 2020
- When searching the ascension number P59594 the information that appears is information on function, names and taxonomy, subcell location, pathology and biotech, PTM/Processing, interaction, structure, family and domain, sequence, similar proteins, cross-references, entry information, and miscellaneous
Part 3
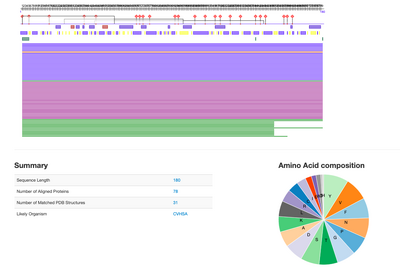
- Using the SARS-CoV sequence from Wan et al. 2020, copy and paste it into PredictProtein server
>2AJF:E|PDBID|CHAIN|SEQUENCE CPFGEVFNATKFPSVYAWERKKISNCVADYSVLYNSTFFSTFKCYGVSATKLNDLCFSNVYADSFVVKGDDVRQIAPGQT GVIADYNYKLPDDFMGCVLAWNTRNIDATSTGNYNYKYRYLRHGKLRPFERDISNVPFSPDGKPCTPPALNCYWPLNDYG FYTTTGIGYQPYRVVVLSFE
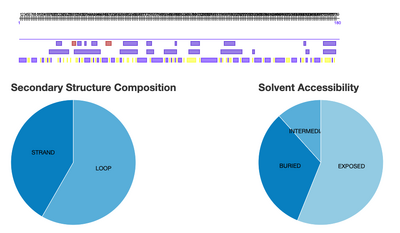
- The types of information available besides amino acid composition and summary (pictured above) are pie graphs depicting secondary structures and solvent accessibility, transmembrane helices, protein disorder and flexibility, disulfide bridges, the effect of point mutations, gene ontology, Terms, Subcellular localization, binding sites, and a literature search
- These relate to UniProt's Information because both focus on the structure of the protein just with different ways of presenting the data, UniProt focuses on structure and interactions, while PredictProtein focused on the individual protein more so
Part 4
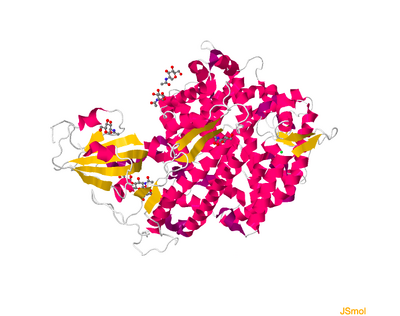
- Using the PDB ID 2AJF, search in the Protein Data Bank for Structure
- The first thing to pop up will be the structure summary, a photo of the 3D structure will be present on the left side of the screen
- Under the Image there will be links to structure, electron density, and ligand interaction, all available in 3D, click on STRUCTURE
- Click on Select Different Viewer, select JSMol
- A picture like below should pop up
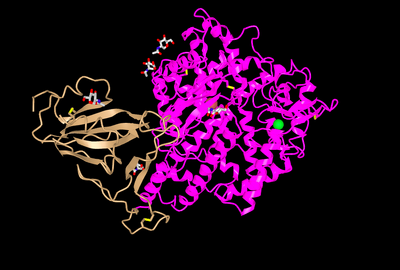
- Another way to get the 3D structure is by using NCBI Structure home page and searching the PDB ID
- To see a full image of the 3D structure click on "full-featured 3D viewer"
- A picture like below should pop up
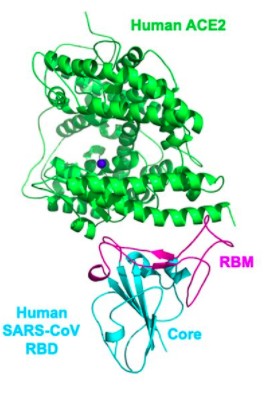
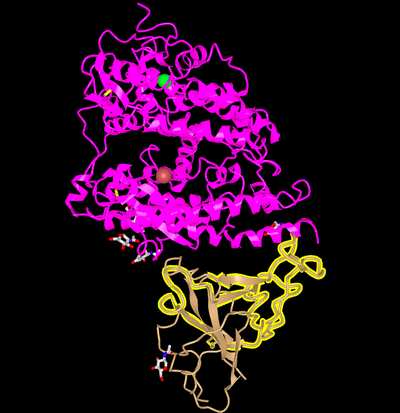
- Orient the Protein similar to Wan et al. 2020
- Wan et al 2020 Picture
- iCn3D Picture
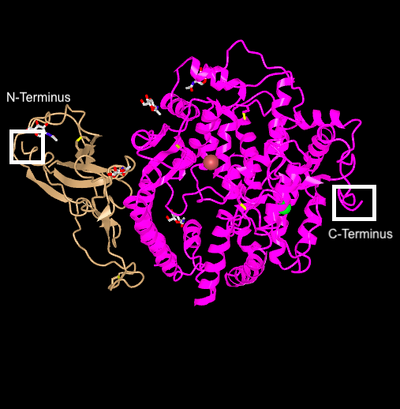
- N and C Terminus are labeled in the picture below
- Yes, the Secondary Structure Predictions match the predictions made by Predict Protein
- Predict Protein Secondary Structure Pie Graph Image Below
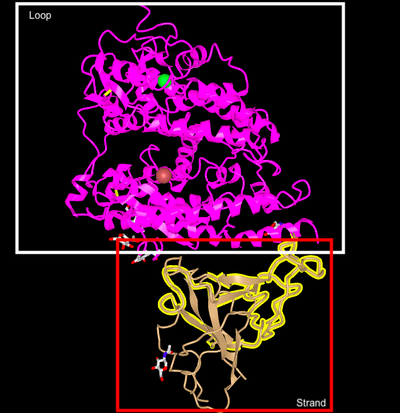
- Labeled Loop and Strand of iCn3D model
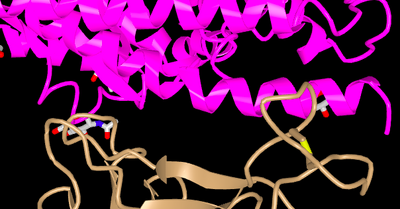
- The paper focused on the binding of ACE2 and SARS-CoV, pictured below is the specific part of the structure that does the binding, with amino acids K,D,E, and M in pink (the ACE2 protein) and T, D, N and F in tan (the SARS-Cov RBM), in the paper the higher-numbered positions were on the RBM and in the 3D model that was made using iCn3D it was switched
Data and Files
- File:First Set of Reading Frames Screen Shot 2020-04-21 at 1.38.44 PM.png
- File:Second Screen Shot 2020-04-21 at 1.38.58 PM.png
- File:MAP Predit Protein Summary.png
- File:MAP JSMOL.png
- File:MAP FUll view 3D.png
- File:MAP Paper copy.png
- File:MAP Replicate of paper.png
- File:MAP Protein NandC.png
- File:MAP Predict Protein .png
- File:Map Labeled loop and strand.png
- File:Highlight amino acids MaP.png
Conclusion
In conclusion, there are different ways of analyzing a 3D structure of a protein from an amino acid sequence. However, it is important when comparing it to academic journals that were published already to correctly orient the models you are analyzing to see correctly if there are similarities or any discrepancies. The 3D structure can give you insight on how amino acids interact with each other. Using the different websites provided you can highlight, color, and focus in on specific parts of the sequence, which allows you to see the most important residues and what they do.
Research Project Proposal
- If species that have not been tested in the Wan et al 2020 paper would fall in the host range of the SAR-CoV2 based on what we know about their ACE2 receptor sequences
- Sequences of Mice, Rat, Civet, Bat, Monkey, Dog, Cat, Bovine, Yeast, Horse, and Chicken, and focusing and comparing the residues that are have found to be important for 2019-nCoV binding to human ACE2
- We plan on using UniProt to obtain those specific sequences, make a phylogenetic tree use the Phylogeny Tree Maker, and then using iCn3D to visualize the interactions that would take place with the SARS-CoV2 protein
References
- Home - ORFfinder - NCBI. (n.d.). Retrieved April 22, 2020, from https://www.ncbi.nlm.nih.gov/orffinder/
- NCBI Protein Domains and Macromolecular Structures. (n.d.). Retrieved April 22, 2020, from https://www.ncbi.nlm.nih.gov/Structure/index.shtml
- OpenWetWare. (2020). BIOL368/S20:Week 13. Retrieved April 22 2020, from https://openwetware.org/wiki/BIOL368/S20:Week_13
- RCSB Protein Data Bank. (n.d.). Homepage. Retrieved April 22, 2020, from https://www.rcsb.org/
- Rost, B., & Yachdav, G. (n.d.). PredictProtein - Protein Sequence Analysis, Prediction of Structural and Functional Features. Retrieved April 22, 2020, from https://ppopen.informatik.tu-muenchen.de/SIB
- Swiss Institute of Bioinformatics. (n.d.). SIB Swiss Institute of Bioinformatics. Retrieved April 22, 2020, from http://web.expasy.org/translate
- UniProtConsortiumEuropean Bioinformatics InstituteProtein Information ResourceSIB Swiss Institute of Bioinformatics. (n.d.). UniProt Consortium. Retrieved April 22, 2020, from http://www.uniprot.org/
- Wan, Y., Shang, J., Graham, R., Baric, R. S., & Li, F. (2020). Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. Journal of virology, 94(7). DOI: 10.1128/JVI.00127-20.
Acknowledgements
- All the information above was obtained from the paper Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. by Wan et al. 2020
- I copied and modified the protocol for Week 13 assignment
- I worked with my homework partners Madeleine B. King and Karina Vescio on developing a research project through text messages
- Madeleine B. King and I asked Kam D. Dahlquist, Ph.D. to work through the new research project with us and asked clarifying questions about the orientation of proteins over email and zoom call on April 21 and 22
- Except for what is noted above, this individual journal entry was completed by me and not copied from another source.
Mpaniag1 (talk) 13:17, 22 April 2020 (PDT)
Classwork
Assignments
Journals
- mpaniag1 Week 2
- mpaniag1 Week 3
- mpaniag1 Week 4
- mpaniag1 Week 5
- mpaniag1 Week 6
- mpaniag1 Week 7
- mpaniag1 Week 8
- mpaniag1 Week 9
- mpaniag1 Week 10
- mpaniag1 Week 11
- mpaniag1 Week 13
- mpaniag1 Week 14