Assignments
Individual Journal Entries
Class Journal Entries
Purpose
The purpose of this week's assignment is to answer the question if rapid progressors and moderate progressors belong in the same category of subjects, since their divergence and diversity had no significant difference.
Combined Methods/Results
Selecting Subjects, Visits, and Clones
- To carry out the experiment, we will only look at the rapid and moderate progressors. We will exclude subject 4 (rapid)and 7(moderate) all together, because they started off with over 1,000 CD4 cells. This could have made the data adjust in a way that is not entirely concise. We will also look at only the first visit and the 2 year mark for the subjects because most of the rapids did not make it past 2 years, so the data will be more uniform between the rapid and moderates. We will also exclude subject 1(rapid) for a similar reason, because they only had three visits which may imbalance the data as well.
- Therefore, the subjects used will be 3,10,11 and 15 for rapid, and 5,6,8,9 and 14 for moderate.
- Looking at the clones for the first visit for the subject (Nucleotide Sequence Data) found on the Week 5 Assignment, the amount we will choose is 3 clones. We decided 3 because subject 6 has the least amount of clones (3), so we want to keep the data uniform as possible. We will also choose them randomly with a calculator so it is non-bias as possible.
- Looking at the clones for the 2 year mark (some of the subjects have the 2 year mark at visit 4 or visit 5), the least number of clones was 6. However we decided to use 3 clones as well, to keep it consistent as well as it would be a lot of sequences to analyze.
- Rapid-
- Subject 3 first visit:4 | 2 year mark(v4): 9
- Subject 10 first visit:7 | 2 year mark(v4): 16
- Subject 11 first visit:7 | 2 year mark(v4): 9
- Subject 15 first visit:12 | 2 year mark(v4): 10
- Moderate-
- Subject 5 first visit:8 | 2 year mark(v4): 7
- Subject 6 first visit:3 | 2 year mark(v5): 9
- Subject 8 first visit:5 | 2 year mark(v4): 6
- Subject 9 first visit:5 | 2 year mark(v5): 9
- Subject 14 first visit:6 | 2 year mark(v5): 7
- To determine the random clones, we used a True Random Generator. The three clones selected are listed below in the table for the first visit. Table was made by Maya. The same goes for the 2 year mark visit.
Randomly Selected Clones from First Visit
| Subject
|
Clone #
|
| 3 (Rapid)
|
1
|
| 2
|
| 4
|
| 5 (Moderate)
|
1
|
| 2
|
| 4
|
| 6 (Moderate)
|
1
|
| 2
|
| 3
|
| 8 (Moderate)
|
2
|
| 4
|
| 5
|
| 9 (Moderate)
|
3
|
| 4
|
| 5
|
| 10 (Rapid)
|
2
|
| 4
|
| 5
|
| 11 (Rapid)
|
4
|
| 5
|
| 7
|
| 14 (Moderate)
|
3
|
| 5
|
| 6
|
| 15 (Rapid)
|
2
|
| 5
|
| 8
|
Randomly Selected Clones from the 2nd Year Visit
| Subject
|
Clone #
|
| 3 (Rapid)
|
1
|
| 6
|
| 8
|
| 5 (Moderate)
|
2
|
| 5
|
| 6
|
| 6 (Moderate)
|
2
|
| 7
|
| 9
|
| 8 (Moderate)
|
1
|
| 3
|
| 4
|
| 9 (Moderate)
|
2
|
| 5
|
| 6
|
| 10 (Rapid)
|
5
|
| 6
|
| 9
|
| 11 (Rapid)
|
1
|
| 6
|
| 9
|
| 14 (Moderate)
|
2
|
| 4
|
| 6
|
| 15 (Rapid)
|
6
|
| 7
|
| 9
|
Making Phylogenetic Trees
- Phylogenetic Trees were made of each subject with their clones from the first visit and clones from the 2nd year visit using the One Click mode on Phylogeny.fr. Subjects 3-9 were made by Maya
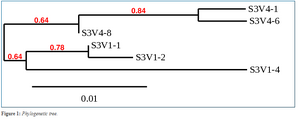
Phylogenetic Tree of Subject 3

Phylogenetic Tree of Subject 5
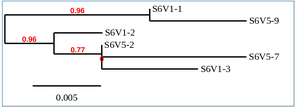
Phylogenetic Tree of Subject 6
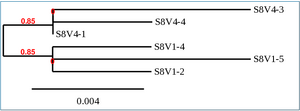
Phylogenetic Tree of Subject 8
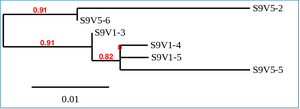
Phylogenetic Tree of Subject 9
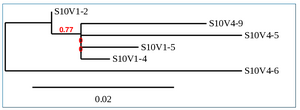
Phylogenetic Tree of Subject 10
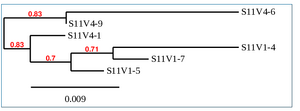
Phylogenetic Tree of Subject 11
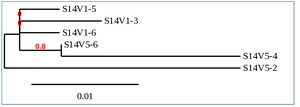
Phylogenetic Tree of Subject 14
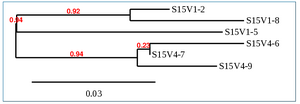
Phylogenetic Tree of Subject 15
Alignment Sequences and Thetas
- Below are the Clustal alignments of each subject with each visit.
- To do this, we added the sequence to Phylogeny.fr and ran the analysis in One Click mode. Then we clicked the Alignment tab and obtained the sequences in Clustal format.
- To calculate the diversity of the clones in each visit, I will use the S statistic by counting the number of position differences between the clones for each subject and calculate θ by using the formula found on the Week 5 assignment page.
- To calculate the number of position differences, I copy and pasted the asterisks into Word for each section and subtracted the number of characters with spaces to ones without spaces.
- To calculate the harmonic sum, I used this calculator. For the number of terms, I used the number of clones.
Subject 3
CLUSTAL FORMAT: SUBJECT 3 FIRST VISIT multiple sequence alignment
S3V1-4 GATGTAGTAATCAGATCCGCCAATTTCACGAACAATGCTAAAACCATACTAGTACAGCTG
S3V1-1 GATGTAGTAATTAGATCCGCCAATTTCTCGGACAATGCTAAAACCATACTAGTACAGCTG
S3V1-2 GATGTAGTAATTAGATCCGCCAATTTCTCGGACAATGCTAAAACCATACTAGTACAGCTG
*********** *************** ** *****************************
S3V1-4 AATGAAACTGTAGTAATGAATTGTACAAGACCCGGCAACAATACAAGAAAAAGAGTAACT
S3V1-1 AATGAAACTGTAGTAATGAATTGTACAAGACCCGGCAACAATACAAGAAAAAGGGTAACT
S3V1-2 AATGAAACTGTAGTAATGAATTGTACAAGACCCGGCAACAATACAAGAAAAAGGGTAACT
***************************************************** ******
S3V1-4 CTAGGACCAGGCAAAGTATACTATACAACAGGACAAATAATAGGAGATATAAGAAAAGCA
S3V1-1 CTAGGACCAGGCAAAGTATACTATACAACAGGACAAATAATAGGAGATATAAGAAAAGCA
S3V1-2 CTAGGACCAGGCAAAGTATACTATACAACAGGACAAATAATAGGAGATATAAGAAAAGCA
************************************************************
S3V1-4 CATTGTAACCTTAGTAGAGCAGATTGGAATAACACTTTAAAAAGGATAGCTATAAAATTA
S3V1-1 CATTGTAACCTTAGTAGAGCAGATTGGAATAACACTTTAAAAAGGATAGCTATAAAATTA
S3V1-2 CATTGTAACCTTAGTAGAGCAGATTGGAATAACACTTTAAAAAGGATAGCTATAAAATTA
************************************************************
S3V1-4 AGAGAACAATTTCAGAATAAAACAATAGTCTTTAATCAATCCTCA
S3V1-1 AGAGAACAATTTCAGAATAAAACAATAGTCTTTAATCAATCCTCA
S3V1-2 AGAGAACAATTTCAGAATAAAACAATAGCCTTTAATCAATCCTCA
**************************** ****************
Subject 3 First Visit
| Subject
|
Number of Clones
|
S
|
Theta
|
| 3
|
3
|
5
|
2.73
|
CLUSTAL FORMAT: SUBJECT 3 2ND YEAR VISIT multiple sequence alignment
S3V4-8 GATGTAGTAATTAGATCCGCCAATTTCGCGGACAATGCTAAAACCATACTAGTACAGCTG
S3V4-1 GATGTAGTAATTAGATCCGCCAATTTCACGGACAATGCTAAAACCATATTAGTACAGCTG
S3V4-6 GATGTAGTAATTAGATCCGCCAATTTCACGGACAATGCTAAAACCATAATAGTACAGCTG
*************************** ******************** ***********
S3V4-8 AATGAAACTGTAGTAATGAATTGTACAAGACCCGGCAACAATACAAGAAAAAGGGTAACT
S3V4-1 AATGAAACTGTAGTAATGAATTGTACAAGACCCGGCAACAATACAAGAAAAAGGGTAACT
S3V4-6 AATGAAACTGTAGTAATGAATTGTACAAGACCCGGCAACAATACAAGAAAAAGGGTAACT
************************************************************
S3V4-8 CTAGGACCAGGCAAAGTATACTATACAACAGGACAAATAATAGGAGATATAAGAAAAGCA
S3V4-1 CTAGGACCAGGCAGAGTATACTATACAACAGGACAAATAATAGGAGATATAAGAAAAGCA
S3V4-6 CTAGGACCAGGCAGAGTATACTATACAACAGGACAAATAATAGGAGATATAAGAAAAGCA
************* **********************************************
S3V4-8 CATTGTAACCTTAGTAGAGCAGGTTGGAATAACACTTTAAAAAGGATAGCTATAAAATTA
S3V4-1 CATTGTAACCTTAGTAGAGCAGGTTGGAATAACACTTTAGAAAGGATAGCTATAAAATTA
S3V4-6 CATTGTAACCTTAGTAGAGCAGGTTGGAATAACACTTTAAAAAGGATAGCTATAAAATTA
*************************************** ********************
S3V4-8 AGAGAACAATTTCAGAATAGAACAATAGTCTTTAATCAATCCTCA
S3V4-1 AGAGAACAATTTCAGAATAGAACAATAGTCTTTAATCAATCCTCA
S3V4-6 AGAGAACAATTTCAGAATAGAACAATAGTCTTTAATCAATCCTCA
*********************************************
Subject 3 2nd Year Visit
| Subject
|
Number of Clones
|
S
|
Theta
|
| 3
|
3
|
4
|
2.18
|
Subject 5
CLUSTAL FORMAT: SUBJECT 5 FIRST VISIT multiple sequence alignment
S5V1-1 GAGGTAGTAATTAGATCCAAAAATTTCTCGGACAATGCTAAAATCATAGTAGTACATCTA
S5V1-4 GAGGTAGTAATTAGATCCAAAAATTTCTCGGACAATGCTAAAATCATAATAGTACATCTA
S5V1-2 GAGGTAGTAATTAGATCCAAAAATTTCTCGGACAATGCTAAAATCATAATAGTACATCTA
************************************************ ***********
S5V1-1 AATGAATCTGTAGAAATTAATTGTACAAGACCCGGCAACAATACAAGAAGAAGTATACAT
S5V1-4 AATGAATCTGTAGAAATTAATTGTACAAGACCCGGCAACAATACAAGAAGAAGTATACAT
S5V1-2 AATGAATCTGTAGAAATTAATTGTACAAGACCCGGCAACAATACAAGAAGAAGTATACAT
************************************************************
S5V1-1 ATAGGACCAGGCAGAGCATTTTATACAACAGGAGACATAATAGGAGATATAAGACAAGCA
S5V1-4 ATAGGACCAGGCAGAGCATTTTATACAACAGGAGACATAATAGGAGATATAAGACAAGCA
S5V1-2 ATAGGACCAGGCAGAGCATTTTATACAACAGGAGACATAATAGGAGATATAAGACAAGCA
************************************************************
S5V1-1 CATTGTAACATTAGTGAAGAAAAATGGAATGAAACTTTAAAAAAGATAGTTATAAAATTA
S5V1-4 CATTGTAACATTAGTGAAGAAAAATGGAATGAAACTTTAAAAAAGATAGTTATAAAATTA
S5V1-2 CATTGTAACATTAGTGAAGAAAAATGGAATGAAACTTTAAAAAAGATAGTTATAAAATTA
************************************************************
S5V1-1 AGAGAACAATTTGGGAATAAAACAATAGTATTTAATCCATCCTCA
S5V1-4 AGAGAACAATTTGGGAATAAAACAATAGTATTTAATTCATCCTCA
S5V1-2 AGAGAACAATTTGGGAATAAAACAATAGTATTTAATCCATCCTCA
************************************ ********
Subject 5 First Visit
| Subject
|
Number of Clones
|
S
|
Theta
|
| 5
|
3
|
2
|
1.09
|
CLUSTAL FORMAT: SUBJECT 5 2ND YEAR VISIT multiple sequence alignment
S5V4-5 GAGGTAGTAATTAGATCCATAAATTTCTCAGACAATGCTAAAATCATAATAGTACATCTA
S5V4-2 GAGGTAGTAATTAGATCTCAAAATCTCTCAGACAATGCTAAAATCATAATAGTACATCTA
S5V4-6 GAGGTAGTAATTAGATCTCAAAATCTCTCAGACAATGCTAAAATCATAATAGTACATCTA
***************** **** ***********************************
S5V4-5 AATGAATCTGTAGAAATTAATTGCACAAGACCCGGCAACAATACAAGAAGAAGTATACAT
S5V4-2 AATGAATCTGTAGAAATTAATTGTACAAGACCCGGCAACAATACAAGAAGAAGTATACAT
S5V4-6 AATGAATCTGTAGAAATTAATTGTACAAGACCCGGCAACAATACAAGAAGAAGTATACAT
*********************** ************************************
S5V4-5 ATAGGACCAAACAGAGCATTTTATACAACAGGAGACATAATAGGAGATATAAGACAAGCA
S5V4-2 ATAGGGCCAAACAGAGCATTTTATACAACAGGAGACATAATAGGAGATATAAGACAAGCG
S5V4-6 ATAGGGCCAAACAGAGCATTTTATACAACAGGAGACATAATAGGAGATATAAGACAAGCA
***** *****************************************************
S5V4-5 CATTGTAACATTAGTGAAGAAAAATGGAATGAAACCTTAAAAAAGATAGTTATAAAATTA
S5V4-2 CATTGTAACATTAGTGAAGAAAAATGGAATGAAACTTTAAAAAAGATAGCTATAAAATTA
S5V4-6 CATTGTAACATTAGTGAAGAAAAATGGAATGAAACTTTAAAAAAGATAGCTATAAAATTA
*********************************** ************* **********
S5V4-5 AGAGAACAATTTAAGAATAAAACAATAGTATTTAAGTCATCCTCA
S5V4-2 AGAGAACAATTTGAGAATAAAACAATAGTATTTAATTCATCCTCA
S5V4-6 AGAGAACAATTTGAGAATAAAACAATAGTATTTAATTCATCCTCA
************ ********************** *********
Subject 5 2nd Year Visit
| Subject
|
Number of Clones
|
S
|
Theta
|
| 5
|
3
|
9
|
4.91
|
Subject 6
CLUSTAL FORMAT: SUBJECT 6 FIRST VISIT multiple sequence alignment
S6V1-1 GAAGTAGTAATTAGATCCGCCAATCTCACGGACAATGCTAAAATCATAATAGTGCATCAG
S6V1-3 GAAGTAGTAATTAGATCCGCCAATCTCACGGACAATGCTAAAATCATAATAGTGCATCTG
S6V1-2 GAAGTAGTAATTAGATCCGCCAATCACACGGACAATGCTAAAATCATAATAGTGCATCAG
************************* ******************************** *
S6V1-1 AATGAATCTGTAGAAATGAATTGTACAAGACCCAACAACAATACAAGAAAAGGTATACAT
S6V1-3 AATGAATCTGTAGAAATGAATTGTACAAGACCCAACAACAATACAAGAAAAGGTATACAT
S6V1-2 AATGAATCTGTAGAAATGAATTGTACAAGACCCAACAACAATACAAGAAAAGGTATACAT
************************************************************
S6V1-1 ATAGGACCAGGCAGAGCATTTTATGCAACAGGAGAAATAATAGGAGATATAAGACAAGCA
S6V1-3 ATAGGACCAGGCAGAGCATTTTATGCAACAGGAGAAATAATAGGAGATATAAGACAAGCA
S6V1-2 ATAGGACCAGGCAGAGCATTTTATGCAACAGGAGAAATAATAGGAGATATAAGACAAGCA
************************************************************
S6V1-1 CATTGTAACCTTAGTAGAGCACAATGGAATGCACATTTAAAAAGGATAGCTATAAAATTA
S6V1-3 CATTGTAACCTTAGTAGAGCACAATGGAATGACACTTTAAAAAGGATAGCTATAAAATTA
S6V1-2 CATTGTAACCTTAGTAGAGCACAATGGAATGACACTTTAAAAAGGATAGCTATAAAATTA
******************************* *************************
S6V1-1 AGAGAACAATTTAAGAATAAAACAATAGTCTTTAATCAATCCTCA
S6V1-3 AGAGAAGTATTTAAGAATAAAACAATAGTCTTTAATCAATCCTCA
S6V1-2 AGAGAACAATTTAAGAATAAAACAATAGTCTTTAATCAATCCTCA
****** *************************************
Subject 6 First Visit
| Subject
|
Number of Clones
|
S
|
Theta
|
| 6
|
3
|
8
|
4.36
|
CLUSTAL FORMAT: SUBJECT 6 2ND YEAR VISIT multiple sequence alignment
S6V5-9 GAAGTAGTAATTAGATCCGCCAATCTCACGGACAATGCTAAAATCATAATAGTGCATCAG
S6V5-2 GAAGTAGTAATTAGATCCGCCAATCTCACGGACAATGCTAAAATCATAATAGTGCATCTG
S6V5-7 GAAGTAGTAATTAGATCCGCCAATCTCACGGACAATGCTAAAATCATAATAGTGCATCTG
********************************************************** *
S6V5-9 AATGAACTTGTAGAAATGAATTGTACAAGACCCAACAACAATACAAGAAAAGGTATACAT
S6V5-2 AATGAATCTGTAGAAATGAATTGTACAAGACCCAACAACAATACAAGAAAAGGTATACAT
S6V5-7 AATGAATCTGTAGAAATGAATTGTACAAGACCCAACAACAATACAAGAAAAGGTATACAT
****** ****************************************************
S6V5-9 ATAGGACCAGGCAGAGCATTTTATGCAACAGGAGAAATAATAGGAGATATAAGACAAGCA
S6V5-2 ATAGGACCAGGCAGAGCATTTTATGCAACAGGAGAAATAATAGGAGATATAAGACAAGCA
S6V5-7 ATAGGACCAGGCAGAGCATTTTATGCAACAGGAGATATAATAGGAGATATAAGACAAGCA
*********************************** ************************
S6V5-9 CATTGTAACCTTAGTAGAGCACAATGGAATGCACATTTAAAAAGGATAGCTATAAAATTA
S6V5-2 CATTGTAACCTTAGTAGAGCACAATGGAATGACACTTTAAAAAGGATAGCTATAAAATTA
S6V5-7 CATTGTAACCTTAGTAGAGCACCATGGAATGACACTTTAAAAAGGATAGCTATAAAATTA
********************** ******** *************************
S6V5-9 AGAGAACAATTTAAGAATAAAACAATAGTCTTTAATCAATCCTCA
S6V5-2 AGAGAACAATTTAAGAATAAAACAATAGTCTTTAATCAATCCTCA
S6V5-7 AGAGAACAATTTAAGAATAAAACAATAGCCTTTAATCAATCCTCA
**************************** ****************
Subject 6 2nd Year Visit
| Subject
|
Number of Clones
|
S
|
Theta
|
| 6
|
3
|
9
|
4.91
|
Subject 8
CLUSTAL FORMAT: SUBJECT 8 FIRST VISIT multiple sequence alignment
S8V1-2 GAGGTAGTAATTAGATCCGTCAATCTCACGGACAATGCTAAAGTCATAATAGTACATCTG
S8V1-4 GAGGTAGTAATTAGATCCGTCAATCTCACGGACAATGCTAAAGTCATAATAGTACATCTG
S8V1-5 GAGGTAGTAATTAGATCCGTCAATCTCACGGACAATGCTAAAGTCATAATAGTACATCTG
************************************************************
S8V1-2 AATGAATCTGTAGAGATGAATTGTACAAGGCCCAACAACAATACAAGAAAAAGTATATCT
S8V1-4 AATGAATCTGTAGAGATGAATTGTACAAGACCCAACAACAATACAAGAAAAAGTATATCT
S8V1-5 AATGAATCTGTAGAGATGAATTGTACAAGACCCAACAACAATACAAGAAAAAGTATATCT
***************************** ******************************
S8V1-2 ATAGGACCAGGCAGAGCATTTTATACAACAGGAGAAATAATAGGAGATATAAGACAAGCA
S8V1-4 ATAGGACCAGGCAGAGCATTTTATACAACAGGAGAAATAATAGGAGATATAAGACAAGCA
S8V1-5 ATAGGACCAGGCAGAGCACTTTATACAACAGGAGAAATAATAGGAGATATAAGACAAGCA
****************** *****************************************
S8V1-2 CATTGTAACCTTAGTAGAGCAAAATGGAATGACACTTTAAAAAATATAGCTATAAAATTA
S8V1-4 CATTGTAGCCTTAGTAGAGCAAAATGGAATGACACTTTAAAAAATATAGCTATAAAATTA
S8V1-5 CATTGTAACCTTAGTAGAGCAAAATGGAATGACACTTTAAAAAATATAGCTATAAAATTA
******* ****************************************************
S8V1-2 AGAGAACAATTTAAGAATAAAACAATAGTCTTTAATCAATCCTCA
S8V1-4 AGAGAACAATTTAAGAATAAAACAATAGTCTTTAATCAATCCTCA
S8V1-5 AGAGAACAATTTAAGAATAAAACGATAGTCTTTAATCAATCCTCA
*********************** *********************
Subject 8 First Visit
| Subject
|
Number of Clones
|
S
|
Theta
|
| 8
|
3
|
4
|
2.18
|
CLUSTAL FORMAT: SUBJECT 8 2ND YEAR VISIT multiple sequence alignment
S8V4-3 GAGGTAGTAATTAGATCCGTCAATCTCACGGACAATGCTAAAGTCATAATAGTACATCTG
S8V4-1 GAGGTAGTAATTAGATCCGTCAATCTCACGGACAATGCTAAAGTCATAATAGTACATCTG
S8V4-4 GAGGTAGTAATTAGATCCGTCAATCTCACGGACAATGCTAAAGTCATAATAGTACATCTG
************************************************************
S8V4-3 AAAGAATCTGTAGAGATGAATTGTACAAGACCCAACAACAATACAAGGAAAAGTATATCT
S8V4-1 AAAGAATCTGTAGAGATGAATTGTACAAGACCCAACAACAATACAAGAAAAAGTATATCT
S8V4-4 AAAGAATCTGTAGAGATGAATTGTACAAGACCCAACAACAATACAAGAAAAAGTATATCT
*********************************************** ************
S8V4-3 ATAGGACCAGGCAGAGCATTTTATACAACAGGGGAAATAATAGGAGATATAAGACAAGCA
S8V4-1 ATAGGACCAGGCAGAGCATTTTATACAACAGGAGAAATAATAGGAGATATAAGACAAGCA
S8V4-4 ATAGGACCAGGCAGAGCATTTTATACAACAGGAGAAATAATAGGAGATATAAGACATGCA
******************************** *********************** ***
S8V4-3 CATTGTAACCTTAGTAGAGCAAAATGGAATGACACTTTAAAAAATATAGCTATAAAATTA
S8V4-1 CATTGTAACCTTAGTAGAGCAAAATGGAATGACACTTTAAAAAATATAGCTATAAAATTA
S8V4-4 CATTGTAACCTTAGTAGAGCAAAATGGAATGACACTTTAAAAAATATAGCTATAAAATTA
************************************************************
S8V4-3 AGAGAACAATTTAAGAATAAAACAATAGTCTTTAATCAATCCTCA
S8V4-1 AGAGAACAATTTAAGAATAAAACAATAGTCTTTAATCAATCCTCA
S8V4-4 AGAGAACAATTTAAGAATAAAACAATAGTCTTTAATCAATCCTCA
*********************************************
Subject 8 2nd Year Visit
| Subject
|
Number of Clones
|
S
|
Theta
|
| 8
|
3
|
3
|
1.64
|
Subject 9
CLUSTAL FORMAT: SUBJECT 9 FIRST VISIT multiple sequence alignment
S9V1-3 GAGGTAGTGATTAGATCCGCCAATTTCACAGACAATGCTAAAACCATAATAGTGCAGCTG
S9V1-4 GAGGTAGTGATTAGATCCGCCAATTTCACAGACAATGCTAAAACCATAATAGTGCAGCTG
S9V1-5 GAGGTAGTGATTAGATCCGCCAATTTCACAGACAATGCTAAAACCATAATAGTGCAGCTG
************************************************************
S9V1-3 AAAGAACATGTAGAAATAAATTGTACAAGACCCAACAACAATACAAGAAAAAGTATAAAT
S9V1-4 AAAGAACATGTAGAAATAAATTGCACAAGACCCAACAACAATACAAGAAAAAGTATAAAT
S9V1-5 AAAGAACATGTAGAAATAAATTGTACAAGACCCAACAACAATACAAGAAAAAGTATAAAT
*********************** ************************************
S9V1-3 ATAGGACCAGGGAGAGCATTTTATGCAACAGGAACAATAATAGGAGATATAAGACAAGCA
S9V1-4 ATAGGACCAGGGAGAGCATTTTATGCAACAGGAACAATAATAGGAGATATAAGACAAGCA
S9V1-5 ATAGGACCAGGGAGAGCATTTTATGCAACAGGAACAATAATAGGAGATATAAGACAAGCA
************************************************************
S9V1-3 CATTGTAACATTAGTGGAGCAAAATGGAATGACACCTTAAAACAGATAGTTGAAAAATTA
S9V1-4 CATTGTAACATTAGTGGAGCAAAATGGAATGACACCTTAAAACAGATAGTTGAAAAATTA
S9V1-5 CATTGTAACATTAGTGGAGCAAAATGGAAGGACACCTTAAAACAGATAGTTGAAAAATTA
***************************** ******************************
S9V1-3 AAAGAACAATTTGAGAATAAAACAATAGTCTTTAATCACTCCTCA
S9V1-4 AGAGAACAATTTGAGAATAAAACAATAGTCTTTAATCACTCCTCA
S9V1-5 AGAGAACAATTTGAGAATAAAACAATAGTCTTTAATCACTCCTCA
* *******************************************
Subject 9 First Visit
| Subject
|
Number of Clones
|
S
|
Theta
|
| 9
|
3
|
3
|
1.64
|
CLUSTAL FORMAT: SUBJECT 9 2ND YEAR VISIT multiple sequence alignment
S9V5-5 GAGGTAGTGATTAGATCCGCCAATTTCACAGACAATGCTAAAACCATAATAGTACAGCTG
S9V5-2 GAGGTAGTGGTTAGATCCGCCAATTTCACAGACAATGCTAAAACCATAATAGTACAGCTG
S9V5-6 GAGGTAGTGATTAGATCCGCCAATTTCACAGACAATGCTAAAACCATAATAGTACAGCTG
********* **************************************************
S9V5-5 AAAGAACATGTAGAAATAAATTGTACAAGACCCAACAACAATACAAGAAAAAGTATAAAT
S9V5-2 AAAGAACATGTAGAAATAAATTGTACAAGACCCAACAACAATACAAGAAAAAGTATAAAT
S9V5-6 AAAGAACATGTAGAAATAAATTGTACAAGACCCAACAACAATACAAGAAAAAGTATAAAT
************************************************************
S9V5-5 ATAGGACCAGGGAGAGCATTTTATGCAACAGGAACAATAATAGGAGATATAAGACAAGCA
S9V5-2 ATAGGACCAGGGAGAGCATTTTATGCAACAGGAGAAATAATAGGAGATATAAGACAAGCA
S9V5-6 ATAGGACCAGGGAGAGCATTTTATGCAACAGGAGCAATAATAGGAGATATAAGGCAAGCA
********************************* ****************** ******
S9V5-5 CATTGTAACATTAGTGGAGTAAAATGGAATGACACCTTAAAACAGATAGCTGAAAAATTA
S9V5-2 CATTGTAACGTTAGTGGAGCAAAATGGCATAACACCTTAAAACAGATAGTTGAAAAATTA
S9V5-6 CATTGTAACATTAGTGGAGCAAAATGGAATAACACCTTAAAACAGATAGTTGAAAAATTA
********* ********* ******* ** ****************** **********
S9V5-5 AGAGAACAATTTCAGAATAAAACAATAGTCTTTAATCACTCCTCA
S9V5-2 AAAGAACAATTTAAGAATAAAACAATAGTCTTTAATCACTCCTCA
S9V5-6 AAAGAACAATTTAAGAATAAAACAATAGTCTTTAATCACTCCTCA
* ********** ********************************
Subject 9 2nd Year Visit
| Subject
|
Number of Clones
|
S
|
Theta
|
| 9
|
3
|
11
|
6.00
|
Subject 10
CLUSTAL FORMAT: SUBJECT 10 FIRST VISIT multiple sequence alignment
S10V1-2 GAGGTAGTAATTAGATCTGAAAATTTCACGGACAATGCTAAAACCATAATAGTACAGCTG
S10V1-4 GAGGTAGTAATTAGATCTGAAAATTTCACGGACAATGCTAAAACCATAATAATACAGCTG
S10V1-5 GAGGTAGTAATTAGATCTGAAAATTTCACGGACAATGCTAAAACCATAATAGTACAGCTG
*************************************************** ********
S10V1-2 AATAAATCTGTAGAAATTAATTGCACAAGACCCAACAACAATACAAGAAGAAGTATAAAT
S10V1-4 AATAAATCTGTAGAAATTAATTGCACAAGACCCAACAACAATACAAGAAGAAGTATAAAT
S10V1-5 AATAAATCTGTAGAAATTAATCGCACAAGACCCAACAACAATACAAGAAGAAGTATAAAT
********************* **************************************
S10V1-2 ATGGGACCAGGGAGAGCATTATATACAACAGGAGAAATAATAGGAGATATAAGGCAAGCA
S10V1-4 ATGGGACCAGGGAGAGCATTTTATACAACAGGAGAAATAATAGGAGATATAAGGCAAGCA
S10V1-5 ATGGGGCCAGGGAGAGCATTTTATACAACAGGAGAAATAATAGGAGATATAAGGCAAGCA
***** ************** ***************************************
S10V1-2 CATTGTAACCTTAGTAGAACAAAATGGAATGACACTTTAAAACAGGTAGTTGACAAATTA
S10V1-4 CATTGTAACCTTAGTAGAACAAAATGGAATGACACTTTAAAACAGGTAGTTGACAAATTA
S10V1-5 CATTGTAACCTTAGTAGAACAAAATGGAATGACACTTTAAAACAGGTAGTTGACAAATTA
************************************************************
S10V1-2 AGAGAACAATTTAGGAATAAAACAATAATCTTTAATCAATCCTCA
S10V1-4 AGAGAACAATTTAGGAATAAAACAATAATCTTTAATCAATCCTCA
S10V1-5 AGAGAACAATTTAGGAATAAAACAATAATCTTTAATCAATCCTCA
*********************************************
Subject 10 First Visit
| Subject
|
Number of Clones
|
S
|
Theta
|
| 10
|
3
|
4
|
2.18
|
CLUSTAL FORMAT: SUBJECT 10 2ND YEAR VISIT multiple sequence alignment
S10V4-5 GAGGTAGTAATTAGATCTGAAAATTTCACGGACAATGCTAAAACCATAATAGTACAGCTG
S10V4-6 GAGGTAGTAATTAGATCTGAAAATTTCACGGACAATGCTAAAACCATAATAGTACAGCTG
S10V4-9 GAGGTAGTAATTAGATCTGAAAATTTCACGGACAATGCTAAAACCATAATAGTACAGCTG
************************************************************
S10V4-5 AATGAATCTGTAGAAATTAATTGCACAAGACCCAACAACAATACAAGAAGAAGCATACAT
S10V4-6 AATAAAGCTGTAGAAATTAATTGCACAAGACCCAACAACAACACAAGAAGAAGAATAAGT
S10V4-9 AATAAATCTGTAGAAATTAATTGCACAAGACCCAACAACAATACAAGAAGAAGTATAAAT
*** ** ********************************** *********** *** *
S10V4-5 ATGGGACCAGGGAGAGCATTTTATGCAACAGGAGAAATAATAGGAGATATAAGGCAAGCA
S10V4-6 ATGGGACCAGGGAGAGTATTGTATACAACAGGAGAAATAATAGGAGATATAAGGCAAGCA
S10V4-9 ATGGGACCAGGGAGAGCATTCTATACAACAGGAGACATAATAGGAGATATAAGGCAAGCA
**************** *** *** ********** ************************
S10V4-5 CATTGTAACCTTAGTAGAACAAAATGGAATGACACTTTAAAACAGGTAGTTGACAAATTA
S10V4-6 CATTGTAACCTTAGTAGAACAAAATGGAATGACACTTTAAAACAGGTAGTTGACAAATTA
S10V4-9 CATTGTAACCTTAGTAGAACAAAATGGCATGACACTTTAAAACAGGTAGTTGACAAATTA
*************************** ********************************
S10V4-5 AGAGAACAATTTAGGAATAAAACAATAATCTTTAAGCAATCCTCA
S10V4-6 AGGGAACAATTTAGGAATAAAACAATAATCCTTAATCAATCCTCA
S10V4-9 AGGGAACAATTTAGGAATAAAACAATAATCTTTAATCAATCCTCA
** *************************** **** *********
Subject 10 2nd Year Visit
| Subject
|
Number of Clones
|
S
|
Theta
|
| 10
|
3
|
14
|
7.64
|
Subject 11
CLUSTAL FORMAT: SUBJECT 11 FIRST VISIT multiple sequence alignment
S11V1-4 GAGGTAATAATTAGATCTAAGAATTTCTCAAACAATGCTAAAAACATAATAGTACAGCTG
S11V1-5 GAGGTAATAATTAGATCTGAGAATTTCTCAAACAATGCTAAAAACATAATAGTACAGCTG
S11V1-7 GAGGTAATAATTAGATCTAAGAATTTCTCAAACAATGCTAAAAACATAATAGTACAGCTG
****************** *****************************************
S11V1-4 AATGAATCTGTAGTAATTAATTGTACAAGACCCGACAACACTATAAAACAAAGGATAATA
S11V1-5 AATGAATCTGTAGTAATTAATTGTACAAGACCCGACAACACTATAAAACAAAGGATAATA
S11V1-7 AATGAATCTGTAGTAATTAATTGTACAAGACCCGACAACACTATAAAACAAAGGATAATA
************************************************************
S11V1-4 CATATAGGACCAGGGAGACCATTCTATACAACAGGAATAAAAGGAAATATAAGACAAGCA
S11V1-5 CATATAGGACCAGGGAGACCATTCTATACAACAGGAATAAAAGGAAATATAAGACAAGCA
S11V1-7 CATATAGGACCAGGGAGACCATTCTATACAACAGGAATAAAAGGAAATATAAGACAAGCA
************************************************************
S11V1-4 CATTGTAACGTTAGTGAGGGACAATGGAATAAAACTTTAGAACAGGTAGTTAGAAAATTA
S11V1-5 CATTGTAACGTTAGTAGAGGACAATGGAATAAAACTTTAGAACAGGTAGTTGGAAAATTA
S11V1-7 CATTGTAACGTTAGTAGAGGACAATGGGATAAAACTTTAGAACAGGTAGTTAGAAAATTA
*************** ********* *********************** ********
S11V1-4 AGAGAACAATATGGACTGAATAAAACAATAGTCTTTAAGCAACCCATA
S11V1-5 AGAGAACAATATGGACTGAATAAAACAATAGTCTTTAAGCAACCCATA
S11V1-7 AGAGAACAATATGGACTGAATAAAACAATAGTCTTTAAGCAACCCATA
************************************************
Subject 11 First Visit
| Subject
|
Number of Clones
|
S
|
Theta
|
| 11
|
3
|
6
|
3.27
|
CLUSTAL FORMAT: SUBJECT 11 2ND YEAR VISIT multiple sequence alignment
S11V4-6 GAGGTAATAATTAGATCTGAGAATTTCTCAAACAATGCTAAAAACATAATAGTACAGCTG
S11V4-1 GAGGTAATAATTAGATCTGAGAATTTCTCAAACAATGCTAAAAACATAATAGTACAGCTG
S11V4-9 GAGGTAATAATTAGATCTGAGAATTTCTCAAACAATGCTAAAAACATAATAGTACAGCTG
************************************************************
S11V4-6 AATGAATCTGTAGTAATTAATTGTACAAGACCCGACTACACTATAAAACAAAGGATAATA
S11V4-1 AATGAATCTGTAGTAATTAATTGTACAAGACCCGACAACACTATAAAACAAAGGATAATA
S11V4-9 AATGAATCTGTAGTAATTAATTGTACAAGACCCGACTACACTATAAAACAAAGGATAATA
************************************ ***********************
S11V4-6 CATATAGGACCAGGGAGACCATTCTATACAACAGGAATAAAAGGAAATATAAGACAAGCA
S11V4-1 CATATAGGACCAGGGAGACCATTCTATACAACAGGAATAAAAGGAGATATAAGACAAGCA
S11V4-9 CATATAGGACCAGGGAGACCATTCTATACAACAGGAATAAAAGGAAATATAAGACAAGCA
********************************************* **************
S11V4-6 CATTGTAACGTTAGTAGAGGACAATGGAATAAAACTTTAGAACAGGTAGTTATAAAATTA
S11V4-1 CATTGTAACGTTAGTAGAGGACAATGGAATAAAACTTTAGAACAGGTAGTTAGAAAATTA
S11V4-9 CATTGTAACGTTAGTAGAGGACAATGGAATAAAACTTTAGAACAGGTAGTTAAAAAATTA
**************************************************** *******
S11V4-6 AGAGAACAATATGGGCCTAATAAAACAATAGTCTTTAAGCAACCCATA
S11V4-1 AGAGAACAATATGGACTAAATAAAACAATAGTCTTTAAGCAACCCATA
S11V4-9 AGAGAACAATATGGACTAAATAAAACAATAGTCTTTAAGCAACCCATA
************** * ******************************
Subject 11 2nd Year Visit
| Subject
|
Number of Clones
|
S
|
Theta
|
| 11
|
3
|
6
|
3.27
|
Subject 14
CLUSTAL FORMAT: SUBJECT 14 FIRST VISIT multiple sequence alignment
S14V1-5 GAGGTAGTAATTAGATCTGAAAATTTCACGAACAATGCTAAAATCATAATAGTACAGCTG
S14V1-6 GAGGTAGTAGTTAGATCTGAAAATTTCACGAACAATGCTAAAATCATAATAGTACAGCTG
S14V1-3 GAGGTAGTAATTAGATCTGAAAATTTCACGAACAATGCTAAAATCATAATAGTACAGCTG
********* **************************************************
S14V1-5 AATGAATCTGTAGAAATTAATTGTACAAGACCCAACAACAATACAAGAAGAAGTATAAAT
S14V1-6 AATGAATCTGTAGAAATTAATTGTACAAGACCCAACAACAATACAAGAAGAAGTATAAAT
S14V1-3 AATGAATCTGTAGAAATTAATTGTACAAGACCCAACAACAATACAAGAAGAAGTATAAAT
************************************************************
S14V1-5 ATAGGACCAGGGAGAGCATTTTATGCAACAGGAGATATAATAGGAGATATAAGGCAAGCA
S14V1-6 ATAGGACCAGGGAGAGCATTTTATGCAACAGGAGATATAATAGGAGATATAAGGCAAGCA
S14V1-3 ATAGGACCAGGGAGAGCATTTTATGCAACAGGAGATATAATAGGAGATATAAGGCAAGCA
************************************************************
S14V1-5 CATTGTAACCTTAGTAGAGCAAAATGGAATGACGCTTTAAAACAGATAGTTTACAAATTA
S14V1-6 CATTGTAACCTTAGTAGAGCAAAATGGAATGACACTTTAAAACAGATAGTTTACAAATTA
S14V1-3 CATTGTAACCTTAGTAGGGCAAAATGGAATGACACTTTAAAACAGACAGTTTACAAATTA
***************** *************** ************ *************
S14V1-5 AGAGAACAATTTGGGAATAATAAAACAATAATCTTTAATCAATCCTCA
S14V1-6 AGAGAACAATTTGGGAATAATAAAACAATAATCTTTAATCAATCCTCA
S14V1-3 AGAGAACAATTTGGGAATAATAAAACAATAATCTTTAATCAATCCTCA
************************************************
Subject 14 First Visit
| Subject
|
Number of Clones
|
S
|
Theta
|
| 14
|
3
|
4
|
2.18
|
CLUSTAL FORMAT: SUBJECT 14 2ND YEAR VISIT multiple sequence alignment
S14V5-2 GAGGTAGTAATTAGATCTGAAAATTTCACGAACAATGCTAAAATCATAATAGTACAGCTG
S14V5-4 GAGGTAGTAATTAGATCTGAAAATTTCACGAACAATGCTAAAATCATAACAGTACAGCTG
S14V5-6 GAGGTAGTAATTAGATCTGAAAATTTCACGAACAATGCTAAAATCATAATAGTACAGCTG
************************************************* **********
S14V5-2 AATGAATCTGTAGAAATTAATTGTACAAGACCCAACAACAATACAAGAAAAAGTATACAT
S14V5-4 AATGAATCTGTAGAAATTAATTGTACAAGACCCAACAACAATACAAGAAAAAGTATAAAT
S14V5-6 AATGAATCTGTAGAAATTAATTGTACAAGACCCAACAACAATACAAGAAGAAGTATAAAT
************************************************* ******* **
S14V5-2 ATAGGACCAGGGAGAGCATTTTATGCAACAGGAAGAATAATAGGAGATATAAGGCAAGCA
S14V5-4 ATAGGACCAGGGAGAGCGTTGTATGCAACAGGAGCTATAATAGGAGATATAAGGCAAGCA
S14V5-6 ATAGGACCAGGGAGAGCATTTTATGCAACAGGAGCTATAATAGGAGATATAAGGCAAGCA
***************** ** ************ ************************
S14V5-2 CATTGTAACCTTAGTAGAGCAAAATGGAATGACACTTTAAAACAGATAGTTTACAAATTA
S14V5-4 CATTGTAACCTTAGTAGAGCAAAATGGAATGACACTTTAAAACAGATAGTTTACAAATTA
S14V5-6 CATTGTAACCTTAGTAGAGCAAAATGGAATGACACTTTAAAACAGATAGTTTACAAATTA
************************************************************
S14V5-2 AGAGAACAATTTGGGAATAATAAAACAATAATCTTTAATCAATCCTCA
S14V5-4 AGAGAACAATTTGGGAATAATAAAACAATAATCTTTAATCAATCCTCA
S14V5-6 AGAGAACAATTTGGGAATAATAAAACAATAATCTTTAATCAATCCTCA
************************************************
Subject 14 2nd Year Visit
| Subject
|
Number of Clones
|
S
|
Theta
|
| 14
|
3
|
8
|
4.36
|
Subject 15
CLUSTAL FORMAT: SUBJECT 15 FIRST VISIT multiple sequence alignment
S15V1-5 GAGGTAGTAATTAGATCTGAAAATTTCACGAACAATGCTAAGATCATAATAGTACATCTG
S15V1-2 GGGGTAGTAATTAGATCTGAAAATTTCACGAACAATGCTAAAATCATAATAGTACAGCTG
S15V1-8 GGGGTAGTAATTAGATCTGAAAATTTCACGAACAATGCTAAAATCATAATAGTACAGCTG
* *************************************** ************** ***
S15V1-5 AATAAATCTGTAGAAATTAATTGTATAAGACCCAACAACAATACAAGAAGAAGGATACCT
S15V1-2 AAGGAAGCTGTAAGAATTAATTGTATAAGACCCAACAACAATACAAGAAGAAGTATACAT
S15V1-8 AAGGAAGCTGTAAGAATTAATTGTATAAGACCCAACAACAATACAAGAAGAAGTATACCT
** ** ***** *************************************** **** *
S15V1-5 ATAGGACCAGGGAGCGCATTTTATACAACAGGCATAATAGGAGATATAAGGCAAGCACAT
S15V1-2 ATAGGACCAGGGAAAACATTTTATACAGGAGACATAATAGGAGATATAAGGCAAGCACAT
S15V1-8 ATGGGACCAGGGAAAGCATTTTATACAGGAGAGATAATAGGAGATATAAGGCAAGCACAT
** ********** *********** ** ***************************
S15V1-5 TGTAACATTAGTATAACAGAATGGAATAACACTTTAAAACAGATAGTTAACAAATTAAGA
S15V1-2 TGTAACATTAGTAGATCAAATTGGAATAGCACTTTAAAACAGATAGTTAACAAATTAAGA
S15V1-8 TGTAACATTAGTAGATCAAAATGGAATCACACTTTAAAACAGATAGTTAACAAATTAAGA
************* * ** * ****** *******************************
S15V1-5 GAACAATTTGTGAATAAAACAATAGTCTTTAATCAGTCCTCA
S15V1-2 GAACAATTTGTGAATAAAACAATAGTCTTTAATCAGTCCTCA
S15V1-8 GAACAATTTGTGAATAAAACAATAATATTTAATCAATCCTCA
************************ * ******** ******
Subject 15 First Visit
| Subject
|
Number of Clones
|
S
|
Theta
|
| 15
|
3
|
27
|
14.7
|
CLUSTAL FORMAT: SUBJECT 15 2ND YEAR VISIT multiple sequence alignment
S15V4-6 GAGGTAGTAATTAGATCTGAAAATTTCACGAACAATGCTAAAATCATAATAGTACATCTG
S15V4-9 GAGGTAGTAATTAGATCTGAAAATTTCACGAACAATGCTAAAATCATAATAGTACATCTG
S15V4-7 GAGGTAGTAATTAGATCTGAAAATTTCACGAACAATGCTAAAATCATAATAGTACATCTG
************************************************************
S15V4-6 AATGAATCTGTAGTAATTAATTGTACAAGACCCAACAACAATACAAGAAGAAAGATACAT
S15V4-9 AATGAATCTGTAGTAATTAATTGTACAAGACCCAACAACAATACAAGAAGAGGGATACAT
S15V4-7 AATGAATCTGTAGTAATTAATTGTACAAGACCCAACAACAATACAAGAAGAAAGATACAT
*************************************************** *******
S15V4-6 ATAGGACCAGGGAAAACATTTTATACAGGAGACATAATAGGAAATATAAGGCAAGCACAT
S15V4-9 ATAGGACCAGGGAAAACATTTTATACAGGAGACATAATAGGAAATATAAGGCAAGCACAT
S15V4-7 ATAGGACCAGGGAAAACATTTTATACAGGAGACATAATAGGAAATATAAGGCAAGCACAT
************************************************************
S15V4-6 TGTAACATTAGTGAGTCAAAATGGAATAACACTTTAAAACAGATAGTTAACAAATTAACA
S15V4-9 TGTAACATTAGGGGTTCAAAATGTAATAACACTTTAAAACAGATAGTTAACAAATTAAGA
S15V4-7 TGTAACATTAGTGGTTCAAAATGGAATAACACTTTAAAACAGATAGTTAACAAATTAAGA
*********** * ******** ********************************** *
S15V4-6 GAACAATTGGAGAATAAAACAATAATCTTTAATCAATCCTCA
S15V4-9 GAACAATTTGGGAATAAAACAATAATCGTTAATCAATCCTCA
S15V4-7 GAACAATTTGGGAATAAAACAATAGTCTTTAATCAATCCTCA
******** * ************* ** **************
Subject 15 2nd Year Visit
| Subject
|
Number of Clones
|
S
|
Theta
|
| 15
|
3
|
11
|
6.00
|
Comparing Theta between First Visit and Two Year Visit
- Maya made this table comparing the thetas between the first visit and the 2nd year visit
| Subject
|
Theta Initial
|
Theta Final
|
| 3
|
2.73
|
2.18
|
| 5
|
1.09
|
4.91
|
| 6
|
4.36
|
4.91
|
| 8
|
2.18
|
1.64
|
| 9
|
1.64
|
6.00
|
| 10
|
2.18
|
7.64
|
| 11
|
3.27
|
3.27
|
| 14
|
2.18
|
4.36
|
| 15
|
14.7
|
6.00
|
Statistic Tests
- To determine the significance between the two groups, a F-test was performed as well as a T-test. Instructions on how to do this in Excel were obtained from t-test protocol and F-test protocol
- The data used to carry out the tests are shown below: the theta differences from the 2nd year visit and first visit separated into rapid and moderate progressors.
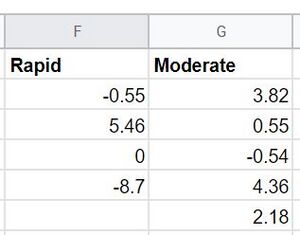
- Before performing a T-test, a F-test must be performed first to test the null hypothesis that the variances of the two populations are unequal.
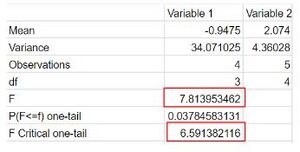
- To do this, the Data Analysis tab was clicked in the Data tab. Then, the F test Two Samples for Variances was selected. The rapid data was inserted in the Variable 1 box, and moderate data in the Variable 2 box.
- If F> F critical one tail, we reject the null hypothesis. We obtained 7.81>6.59 so this is true. Therefore, we reject the null hypothesis. The variances of the rapid and moderate progressors are unequal and we can proceed to do the t-test.
- Now we can perform the T-Test. A T-Test is used to test the null hypothesis that the means of two populations are equal, and in this case, determine if rapid and moderate progressors should be categorized in the same group.
- To do this, the Data analysis tab was clicked in the Data tab. Then, t-test: Two Samples Assuming Unequal Variances was selected. The rapid data was inserted in the Variable 1 box, and moderate data in the Variable 2 box. The Hypothesized Mean Difference was inserted to be 0.
- If t Stat < -t Critical two-tail OR if t-stat > t Critical two-tail, we reject the null hypothesis. However, this is not true, since we obtained: -0.986<-2.776 or -0.986>2.77.
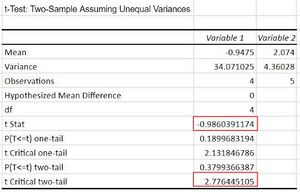
- Therefore, we don't reject the null hypothesis. The observed difference between the sample means (we obtained 0.94,2.074) is not convincing enough to say that the average genetic difference between rapid and moderate differ significantly.
- Therefore, they belong in the same group according to this data.
- By looking at the variances of the two populations, (34,4.36), it can be determined why the p value could be big. There is a 8-fold difference between the moderate and rapid progressors, which leads to there being no statistical difference between rapid and moderates.
- Also, the small sample size could contribute to why the p value was greater than 0.05. If there were more subjects, there would be less bias and the p value could go down.
Data and Files
Phylogenetic trees of all the subjects studied are linked below. Subjects 3-9 were made by Maya. Subjects 10-15 were made myself.
All the statistical data including the theta differences, F-test results, and T-test results are in the zip file below.
Final Project Powerpoint (PDF)
Maya uploaded our presentation as a PDF
Scientific Conclusion
Markham et. al (1998) conducted a study regarding HIV-1 evolution and determined the pattern of divergence and diversity between and within subjects. They placed the subjects into three different categories, one is rapid, the second moderate, and the third non-progressive. However, when they performed their experiment, they concluded that rapid and moderate progressors were not statistically different, even though they were placed in different groups. Our main question was that since rapid and moderate progressors had no significant difference, could be placed in the same group of subjects? After doing some data cleanup and calculating genetic differences between two separate visits, we performed a T-test to determine if the means of the rapid and moderate progressors were significantly different. The t-test showed that the means of the two groups were not significantly different, so rapid and moderate progressors should be in the same group. We also noticed the high variance and small sample size of the results, and concluded that these values could have lead to the rapid and moderate progressors being not significantly different. Further research includes a bigger sample size and more consistent visits in order to get a less bias p value, based on those factors.
Acknowledgments
- I copied and modified the protocol from Week 6 for this assignment.
- I worked with my project partner Maya in and outside of class.
- I communicated with Dr. Dahlquist with questions about statistical analysis
- Maya and I went to Dr. Dahlquist on Wednesday to talk about our powerpoint presentation.
- I used Phylogeny.fr to make my phylogenetic trees and obtain clustal alignments.
- I used the harmonic sum calculator found at Dcode.fr
- I copied and modified the protocol for F-test and T-test from Excel-easy and [1]
- I obtained the sequence data for HIV subjects, visits, and clones from Nucleotide Sequence Data found on the Week 5 Assignment page.
- I used a random number generator to randomize the clones selected from True Random Generator
- I copied and modified the syntax from Maya's Week 6 to make the tables
- Except for what is noted above, this individual journal entry was completed by me and not copied from another source.
Mking44 (talk) 11:18, 24 February 2020 (PST)
References
- OpenWetWare. (2020). BIOL368/S20:Week 6. Retrieved February 24, 2020, from https://openwetware.org/wiki/BIOL368/S20:Week_6
- dCode. (n.d.). Harmonic Number and Series Calculator - Online Software Tool. Retrieved February 24, 2020, from https://www.dcode.fr/harmonic-number
- F-Test in Excel. (n.d.). Retrieved February 26, 2020, from https://www.excel-easy.com/examples/f-test.html
- Haahr, M. (n.d.). True Random Number Service. Retrieved February 24, 2020, from https://www.random.org/
- Nucleotide Sequences. (n.d.). Retrieved February 24, 2020, from http://bioquest.org/bedrock/problem_spaces/hiv/nucleotide_sequences.php
- Phylogeny.fr References
- http://www.phylogeny.fr/
- Dereeper A.*, Guignon V.*, Blanc G., Audic S., Buffet S., Chevenet F., Dufayard J.F., Guindon S., Lefort V., Lescot M., Claverie J.M., Gascuel O. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008 Jul 1;36(Web Server issue):W465-9. Epub 2008 Apr 19 (PubMed)
- Dereeper A., Audic S., Claverie J.M., Blanc G. BLAST-EXPLORER helps you building datasets for phylogenetic analysis. BMC Evol Biol. 2010 Jan 12;10:8. (PubMed)
- t-Test in Excel. (n.d.). Retrieved February 24, 2020, from https://www.excel-easy.com/examples/t-test.html











