Mking44 Week 14
Assignments
Individual Journal Entries
Class Journal Entries
Purpose
The purpose of this project was to investigate the ACE2 ortholog by comparing other species not found in the Wan et al. (2020) paper and determining sequence-structure-function relationships when binding to 2019n-CoV
Combined Methods/Results
Obtaining ACE2 orthologs
- 19 species that have ACE2 receptor were chosen from Uniprot.
- Some species were in Wan et. al (2020): Civet, Bat, Mouse, Rat, Pig, and Orangutan
- New species were added: Platypus, Opossum, Elephant, Sperm Whale, Horse, Dog, Red Fox, Chicken, Turtle, Chameleon, and Zebrafish.
- ACE2 receptor was inputted into database, and sequences were obtained in FASTA format.
Phylogeny and Multiple Sequence Alignment
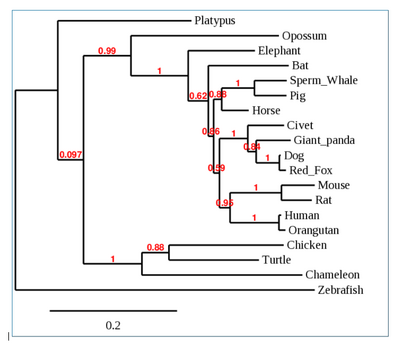
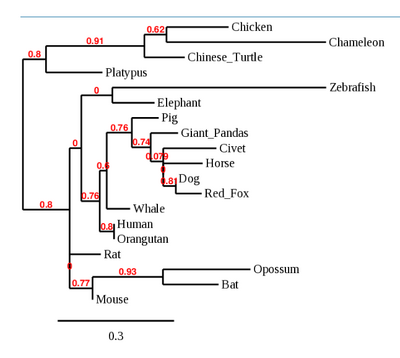
- Phylogeny.fr was used in order to see phylogenetic trees of the 19 species with ACE2 orthologs
- Phylogenetic Tree was generated of the entire ACE2 receptor sequence as seen below:
- No distinct pattern is observed, however, some groups were predicted to be classified together. (i.e. Human and Orangutan, Mouse and Rat, Platypus and Zebrafish as outliers).
- Another Phylogenetic Tree was generated of just the binding region of ACE2 receptor as seen below:
- No distinct pattern was observed for binding region of ACE2 receptor.
- Multiple Sequence Alignment of the sequences were conducted in CLUSTAL format
CLUSTAL FORMAT: MUSCLE (3.8) multiple sequence alignment
tr|Zebrafish| QTVEDRAREFLNKFDEEASDIMYQYTLASWAYN
tr|Chameleon| DVTQQAAE-FLLQFNINAENRSYESSLASWDYN
tr|Chicken| DVTQE-AQTFLAEFNVRAEDISYENSLASWNYN
tr|Turtle| DITQEAIN-FLSEFNVQAEDLSYASSLASWNYN
tr|Platypus| K-PEEEARQFLTQFNKQAEDLSYQSSLASWEYN
tr|Opossum| NSIEEDAKTFLDDYNAKAEELSHQSALASWEYN
sp|Mouse| SLTEENAKTFLNNFNQEAEDLSYQSSLASWNYN
sp|Rat| SLIEEKAESFLNKFNQEAEDLSYQSSLASWNYN
tr|Bat| STTEDEAKMFLDKFNTKAEDLSHQSSLASWDYN
tr|Elephant| STTEDLARTFLDTFNQEAEDLSYQSSLASWDYN
sp|Human| STIEEQAKTFLDKFNHEAEDLFYQSSLASWNYN
sp|Orangutan| STIEEQAKTFLDKFNHEAEDLFYQSSLASWNYN
tr|Whale| STTEEQAKTFLQKFDHEAEDLSYQSSLASWNYN
tr|Pig| STTEELAKTFLEKFNLEAEDLAYQSSLASWNYN
tr|Horse| STTEDLAKTFLEKFNSEAEELSHQSSLASWSYN
sp|Civet| STTEELAKTFLETFNYEAQELSYQSSVASWNYN
tr|Panda| STTEDLAETFLEKFNYEAEDLYYQSSLASWNYN
tr|Dog| S-TEDLVKTFLEKFNYEAEELSYQSSLASWNYN
tr|Red Fox| S-TEDLVNTFLEKFNYEAEELSYQSSLASWDYN
:: ** :: *.: : ::*** **
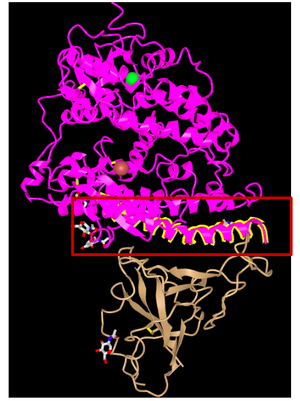
Viewing the 3D structure of ACE2 and 2019n-CoV interaction
- iCn3D viewer was used in order to view the binding interaction
- the Protein Data Bank ID for Wan et al (2020) was used: 2AJF
- As shown below, the first alpha helix at positions 1-20 is highlighted, which is seen to 'touch' or is closest to the SARS-CoV
- Note: in 3D viewer, the alpha helix is position 1-33, but in the paper, it is actually positions 19-51.
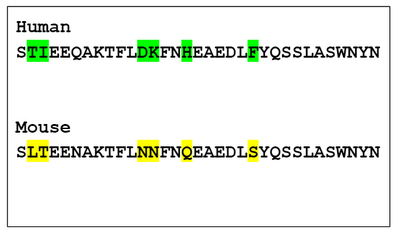
Comparing Mouse and Human ACE2 Ortholog with Alpha Helix
- In Wan et al 2020, they predicted that mouse could not bind to SARS-2 due to residue differences in specific positions in the ACE2 sequence.
- Therefore, knowing that human ACE2 receptor can bind to SARS-2, we wanted to compare human and mouse sequences using the alpha helix position we found in the 3D viewer.
- As seen, human and mouse receptors were compared and were seen to have 6 residue differences that would cause a difference in the interaction.
- The glutamine changed to asparagine was also observed, but this would not make a difference since they both are polar and have similar structures.
Choosing Species to Compare 6 Residues
- Due to limited time, 5 species were selected from phylogenetic tree to compare to human ACE2 ortholog:
- Orangutan (due to similarity with Human), Civet (known to have unfavorable interactions with SARS-CoV), Horse(similar to Civet on tree), Platypus (as an outlier), and Bat(known to carry coronaviruses).
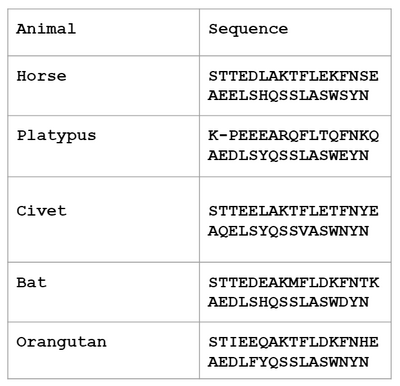
- Alpha helix ACE2 ortholog sequences are shown for the five species chosen.
Horse
- About half of the residues are conserved in Horse
- I changed to a T = nonpolar to polar
- D changed to an E = both negative charged, so not a huge effect
- H changed to an S = positive charged to polar
- F changed to an S = nonpolar to polar
- This can be expected, since it's similarly related to Civet, it is known that SARS binds unfavorably to Civet.
- Horses are known to get sick with certain corona viruses in general.
Platypus
- Platypus had differences in every residue, which can be partially expected since it was the outlier in the phylogenetic tree and in general in the species chosen.
- T deleted = loss of polar amino acid
- I changed to P= both hydrophobic, but proline is a different shape and can disrupt a helix
- D changed to T= negative charged to polar
- K changed to Q= positive charged to polar
- H changed to K= both positive charged, but change of shape
- F changed to S= nonpolar to polar
Civet
- In Wan et al.(2020), it is noted that civets have unfavorable binding interactions with SARS-2, and is also known as the carrier of SARS-1.
- It can be expected there be some similarity, but not entirely
- Only one of the residues were conserved:
- I changed to a T= nonpolar to polar
- D changed to an E= both negatively charged, so not a big effect
- K changed to a T= positive charged to polar
- H changed to a Y= positive charged to polar
- F changed to a S= nonpolar to polar
Bat
- Bats are known to be carriers of coronavirus, so it is predicted that some homology will be seen between the two orthologs.
- About half of the residues are conserved:
- I changed to T= nonpolar to polar
- H changed to T= positive charged to polar
- F changed to S= nonpolar to polar
Orangutan
- It is known that 97-98% DNA is shared between human and orangutan, so we expected the sequences to be very similar
- All of the residues, including the entire alpha helix ended up being identical
- This can be seen where great apes around the world are being put on lock down due to the coronavirus outbreak.
ACE2 Ortholog Summary
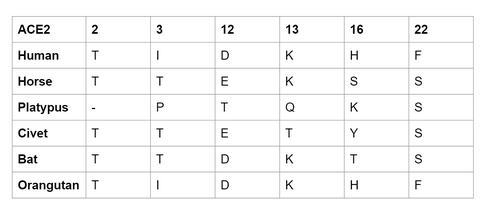
- Residue differences of the 5 species compared to human ortholog can be summarized in the table below:
- For residue 2: All of them have a T, except platypus.
- For residue 3: Primates have I, the rest have T, except platypus. I is nonpolar, T is polar.
- For residue 12: All of them either have a D or E, except for platypus. D and E are in the same group so it doesn't make much of a difference.
- For residue 13: All of them have a lysine, except civet (which binds unfavorably) and platypus.
- This is the lysine 31 that is in the Wan et al. (2020) paper.
- For residue 16: Primates have H. The rest have Y, S, and T. H and Y have similar shapes, T and S are polar but does not have a ring structure.
- For residue 22: Primates have F, the rest have S. S has a polar group, F has a aromatic ring.
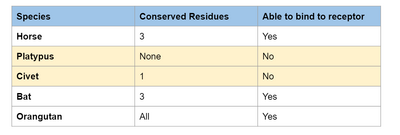
Can These Species Bind to SARS?
- We decided that residues that have half or more of the conserved residues could be predicted to be able to bind to SARS-2.
- Therefore, following this analysis, we predict that horse, bat, and orangutan should be able to bind to the virus.
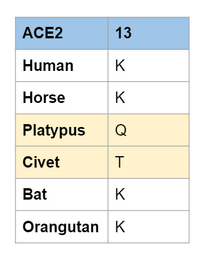
Comparing position 13 to Wan et al. (2020)
- We realized after we did our experiment that position 13 is the same lysine 31 that is discussed in the Wan et al. (2020) paper.
- The numbering is off due to them cutting down the sequence for the 3D structure
- They discuss that Lysine 31 is an important hot spot for SARS-CoV to bind to the host receptor.
- It forms a salt bridge with Glu35 and is buried in a hydrophobic region.
- Since the lysine is changed to a threonine, it cannot form a salt bridge with Glu35 and leads to unfavorable interactions with the virus.
- This supports our hypothesis that civet will not be able to bind well to the virus.
Discussion & Future Research
- Our predictions can give an idea if the virus can bind to a particular ACE2 ortholog
- However, we just looked at the helix that directly contacts the virus.
- Other parts of the ACE2 structure can play a role in binding as well.
- Civets were able to have SARS-CoV-1, but predicted to not bind to SARS-2 due to unfavorable bonds.
- This could give us a hint in what makes the two viruses different.
- It is recorded that several big cats and household cats have positive antibodies for coronavirus.
- Different species of cats ACE2 receptors can be compared with civets in order to find specific differences.
Data and Files
- File:Uniprotfastamking.PNG
- FASTA sequences.zip File:Uniprot FASTA sequences.zip
- File:Phylogenetictreeace2mking.PNG
- File:Bindingregionace2phylotreemking.PNG
- File:3dviewerace2mking.PNG
- File:Humanmouseace2mking.PNG
- File:5speciesmodifiedhorsemking.PNG
- File:Horsemking.PNG
- File:Platypusmking.PNG
- File:Civetmking.PNG
- File:Batmking.PNG
- File:Orangutanmking.PNG
- File:Ace2tablesummarymkingmodified.PNG
- File:Comparingresiduesmking.PNG
- File:Position13mking.PNG
Final Powerpoint PDF
File:Maddykarinamayalizzy1.pdf
Scientific Conclusion
ACE2 is a enzyme found in a wide range of species from mammals to fish, birds, and reptiles. We wanted to see how other species ACE2 ortholog could compare to the human ACE2 ortholog as well as how well it binds to SARS-CoV-2. We concluded that horse, bat, and orangutan ACE2 could all bind to 2019-nCov due to sequence conservation in the alpha helix that directly contacts the virus. An important binding site lysine31 is mentioned in Wan et al. that corresponds to position 13 in the alpha helix. All of the species chosen have the lysine31 except the civet and platypus. Therefore, this is further evident that platypus and civet do not bind favorably to the virus if not at all.
Acknowledgments
- I copied and modified the protocol from Week 14 for this assignment.
- I worked with my homework partners for this week to complete this presentation and obtain data: Maya, Karina, and Lizzy
- I worked with my homework partners in class, texted my groupmates about writing our slides, and met with them on Wednesday April 29th to practice the presentation.
- The basis for this project was built off of Wan et al. (2020) [cited in references]
- Except for what is noted above, this individual journal entry was completed by me and not copied from another source.
Mking44 (talk) 14:29, 28 April 2020 (PDT)
References
- OpenWetWare. (2020). BIOL368/S20:Week 14. Retrieved April 28, 2020, from https://openwetware.org/wiki/BIOL368/S20:Week_14
- Phylogeny.fr References
- http://www.phylogeny.fr/
- Dereeper A.*, Guignon V.*, Blanc G., Audic S., Buffet S., Chevenet F., Dufayard J.F., Guindon S., Lefort V., Lescot M., Claverie J.M., Gascuel O. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008 Jul 1;36(Web Server issue):W465-9. Epub 2008 Apr 19 (PubMed)
- Dereeper A., Audic S., Claverie J.M., Blanc G. BLAST-EXPLORER helps you building datasets for phylogenetic analysis. BMC Evol Biol. 2010 Jan 12;10:8. (PubMed)
- Cn3D. Bethesda (MD): National Library of Medicine (US), National Center for Biotechnology Information; 2020 – [cited 2020 Apr 28]. Available from: http://www.ncbi.nlm.nih.gov/Structure/CN3D/cn3d.shtml
- UniProtKB (2020). National Center for Biotechnology Information (NCBI). Retrieved April 28, 2020, from https://www.uniprot.org
- Wan, Y., Shang, J., Graham, R., Baric, R. S., & Li, F. (2020). Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. Journal of virology, 94(7). DOI: 10.1128/JVI.00127-20.