Mking44 Week 2
Assignments
Individual Journal Entries
Class Journal Entries
Purpose
The purpose of this assignment was use a bio-mathematical model, along with knowledge of the central dogma, to determine how the DNA sequence of a pigment protein gene determines the color of a protein. Then, a true-breeding purple flower will be created to show this understanding.
Class Notes
- differences between blue and yellow protein: position 79 and 80.
- differences between red and white protein: position 7
- differences between blue and green protein: position 83
- differences between yellow and green: position 79, 80, and 83.
- differences between red and blue: position 79.
- differences between white and blue: position 79 and 78.
- differences between white and yellow: position 78, 79, 80
- differences between yellow and red: positions 79 and 80.
Combined Methods and Results
Methods
- Downloaded Aipotu program (link found in Acknowledgements)
- Clicked on the 'Molecular Biology' Tab
- Familiarized myself with the program by following sample experiments in pages 2,4-6.
Specific Tasks
A. What are the differences in the DNA sequences of the alleles you defined in Part I?
- The alleles from the Part 1 Genetics were green (C^G), red (C^R), blue (C^B), yellow (C^Y), and white (C^W and C^w)). It was found through self-crossing that some were incomplete dominance, and white was recessive to everything.
- I found the DNA sequences of the alleles by comparing the sequences of the different alleles found in the organisms:
- For example, I double-clicked the Green-2 Organism in the Greenhouse section to the right
- The upper DNA sequence is shown to be the blue protein, and the lower DNA sequence is shown to be the yellow protein, both combining to get green.
- Compare Tab -> Upper vs. Lower
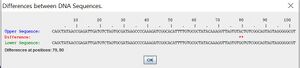
- As displayed in Figure 1, the differences between the blue allele and the yellow allele are above. They are different at positions 79 and 80, with an AC for blue and a GG for yellow.
- It also shows you the differences in the amino acid sequence between two alleles as well.
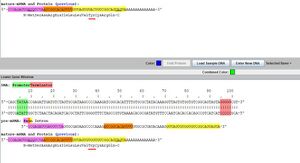
- By looking at the mature-mRNA and protein in Figure 2, it can be shown that tyrosine and tryptophan are different in the blue alle (upper) and yellow allele (lower)
- The same procedure was used for the rest of the alleles by clicking on the different organisms (Green 1, Red and White).
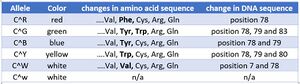
- The rest of the sequences are shown in Table 1.
- It is observed by looking at the organisms, that there are two white alleles. One in the red flower (C^w), and one in the white flower(C^W). The one in the red flower has no promoter, and no mRNA or protein is expressed. The one in the white flower makes a white protein.
B. Do all the white alleles have the same DNA sequence? Are the white alleles the same?
- To answer the question, I have already compared the white alleles found in Table 1.
- No the white alleles do NOT have the same DNA sequence. It is observed that there are differences in position 7 and 78. There is a mutation in the one in the red flower, leading to no transcription or translation. The one in white flower makes a white/colorless protein.
- Therefore, the white alleles are not the same, even though they express the same "color."
C. Which DNA sequences are found in each of the four starting organisms?
- To find the DNA sequences of each four starting organisms, I followed the instructions found in the answer to question A, but just did it for each organism.
- Green-1:
- Upper & Lower(identical):CAGCTATAACCGAGATTGATGTCTAGTGCGATAAGCCCCAAAGATCGGCACATTTTGTGCGCTATACAAAGGTTAGTGTACTGGCGGCAGTAGTAGGGGGCGT
- Green-2:
- Upper: CAGCTATAACCGAGATTGATGTCTAGTGCGATAAGCCCCAAAGATCGGCACATTTTGTGCGCTATACAAAGGTTAGTGTACTGTCGGCAGTAGTAGGGGGCGT
- Lower: CAGCTATAACCGAGATTGATGTCTAGTGCGATAAGCCCCAAAGATCGGCACATTTTGTGCGCTATACAAAGGTTAGTGTGGTGTCGGCAGTAGTAGGGGGCGT
- Red:
- Upper:CAGCTATAACCGAGATTGATGTCTAGTGCGATAAGCCCCAAAGATCGGCACATTTTGTGCGCTATACAAAGGTTAGTGTTCTGTCGGCAGTAGTAGGGGGCGT
- Lower:CAGCTATGACCGAGATTGATGTCTAGTGCGATAAGCCCCAAAGATCGGCACATTTTGTGCGCTATACAAAGGTTAGTGTTCTGTCGGCAGTAGTAGGGGGCGT
- White:
- Upper & Lower(identical):CAGCTATAACCGAGATTGATGTCTAGTGCGATAAGCCCCAAAGATCGGCACATTTTGTGCGCTATACAAAGGTTAGTGGTCTGTCGGCAGTAGTAGGGGGCGT
D. Using this knowledge, construct a pure-breeding purple organism
- My partners and I's first instinct to create a purple organism involved combining a red and a blue gene. However, it is not possible to simply take the two genes and create a flower.
- We decided to try to mutate the yellow protein in the green-2 to a red protein, so it can combine with the blue gene in Green-2.
- By using the Compare tool, yellow and red protein differed at position 7, 79, and 80.
- Therefore, we changed the nucleotide differences by clicking on the position and typing in the desired base (A,T,C,G).
- This made a purple flower.
- However, it was not true breeding, just the red and blue gene combined to create a purple phenotype.
- We then hypothesized we had to combine the two genes somehow to create a true breeding purple one.
- By observing the differences between the red and blue amino acid sequences in Table 1, they have the same sequence, except in the fourth to last position, there is a Phe in red and a Tyr in blue.
- So, we decided to insert a Tyr following the Phe in the red gene sequence.
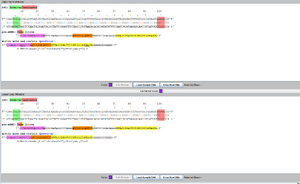
- It worked! (see Figure 3 and 4)
- It was also observed that the order of inserting did not matter.
- For example, you could add a Phe before or after the Tyr, and vise versa with Tyr. And if you matched two pure-breeding purple flowers with different sequences, the end result is still a purple flower.

E. Advanced tasks: How does the DNA sequence of the different alleles explain the effects of mutations you found in part I?
- In part I, the Genetics group (Maya's mutation) discovered that by mutating a pure red flower, it turned into a purple flower. It also was discovered that several amino acids were added to create the flower including Tyr which is found in the blue flower.
F. Try making this protein: MLVKEIAMYRFATHER
- Since the letters represent the amino acids, one can use the genetic code to write a nucleotide sequence that translate to that protein.
- For example: M-> Met-> ATG, L-> Leu->,CTT
- Figure 5 shows the result of making the custom protein
- I did this by editing one of the sequences by typing in the desired nucleotides. (The blue shows the previous sequence)

Data and Files
Scientific Conclusion
The color of a pigment protein isn't simple as just the amino acid sequence. It involves the DNA sequence of that gene that will be transcribed to a mRNA then translated to a mature protein. It was shown that the alleles weren't as simple as dominant and recessive, but involved incomplete dominance, and even a non-transcribing gene found in the red flower. It was also discovered that two flowers of the same color doesn't necessarily mean that the amino acid sequence or even the DNA sequence were the same. For example, there was a Green-1 and Green-2 organism: one with a blue and yellow protein, the other with two green proteins. To create a purple flower, one can just mutate the yellow protein in the Green-2 organism to be red, and red combines with blue protein to create purple. However, it is not true breeding since the two alleles are showing incomplete dominance. To create the purple true-breeding flower, one must compare the blue and red amino acid and DNA sequence and combine the two differences together to create a mutated flower. It doesn't matter if the blue or red protein comes first, as long as the two differences are next to each other.
Acknowledgements
- I worked with my partners Nathan On and Karina Vescio on reading the protocol and answering the questions during classtime.
- I collaborated with Maya outside of lab on Monday to work on syntax and formatting.
- I used Maya's image on her individual journal to help answer part E (since it asks about part 1)
- I copied and modified Aipotu's protocol for the molecular biology section in my laboratory notebook.
- I used the syntax for adding images and and links shown on the Week 1 page.
- I copied and modified the protocol on Week 2 for this assignment
- I read the articles for the class journal assignment found on the Week 2 page
- I looked at Christina's individual journal for help with formatting
- I downloaded and used [Aipotu] program in order to complete this assignment
- Except for what is noted above, this individual journal entry was completed by me and not copied from another source.
Mking44 (talk) 00:07, 28 January 2020 (PST)
References
- OpenWetWare. (2020). BIOL368/S20:Week 2. Retrieved January 27, 2020, from https://openwetware.org/wiki/BIOL368/S20:Week_2
- Aipotu. (2017). Aitopu. Retrieved January 27, 2020, from http://aipotu.umb.edu/
- Aipotu. (2017). Aipotu Part III: Molecular Biology. Retrieved January 27, 2020, from http://aipotu.umb.edu/docs/AipotuIII.pdf
- OpenWetWare. (2020). File:Maya Mutation.png. Retrieved January 27, 2020, from https://openwetware.org/wiki/File:Maya_Mutation.png