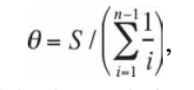Matthew K. Oki Individual Journal 4
Matthew K. Oki I.J. Week 4
Purpose
The purpose of this week's exercise was to deepen our knowledge of Biology Workbench and sequence comparison. We will be needing an understanding of these tools in order to complete our own studies of HIV sequences in the coming weeks.
Methods & Results
- The formal methods were followed from Activity 2 of the Exploring HIV Evolution
Activity 2, Part 1
- Download the provided sequences of subjects 10-15 from BIOL368/F16:Week 4
- Upload these sequences into the Biology Workbench
- Run the multiple sequence alignment for the selected subjects and clones
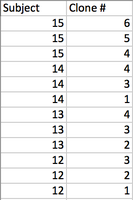
- The subjects & clones below were used:
- The output results in the following uprooted tree:
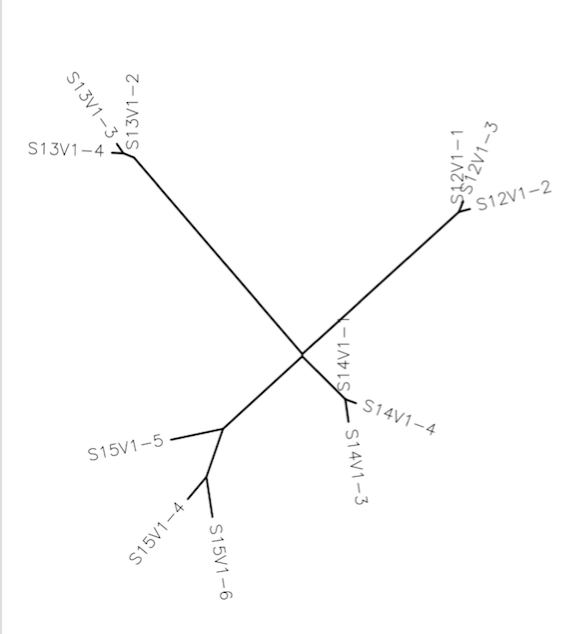
- The clones on the tree seemed to cluster together.
- The clones of subject 15 had a good amount of variance compared to those of the other subjects.
- It appeared as if subjects 15 and 14 were closer to each other (more clustered) than the other two subjects.
- The closer the subjects were to each other in respect to the central connecting point, the more related they were. So, subjects 15 and 14 were fairly related to each other, while the other two (subjects 12 and 13) were not nearly as related.
Activity 2, Part 2
- Three individual subjects were run through multiple sequence alignment
- Each nucleotide difference (S) was denoted by the absence of a star as shown below with Subject 15:
- Theta was then calculated:
- Theta is an estimate of the average pairwise genetic distance
- The equation from Exploring HIV Evolution is given below:
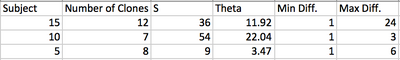
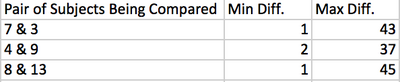
- Min and Max difference were calculated:
- One subject's clones were selected and run through the same multiple sequence alignment
- Click on "Import Alignments"
- Go to the "Alignment Tool" tab
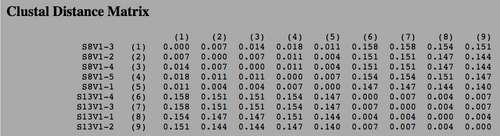
- Run a "Clustaldist" test
- A matrix is produced as shown below:
- The highest and lower numbers in this matrix were multiplied by the length of sequence
- Rounding these numbers to the nearest integer gives us maximum and minimum respectively
- This was performed for all 3 of the subjects picked above with the results in the table below:
- Finally, the min and max were calculated from a comparison of two subjects:
- The table below gives the results of three comparisons:
Data & Files
Scientific Conclusion
Subjects 15, 14, and 12 seemed to be the most related, as they were the closest together on the unrooted tree created in the ClustalW sequence alignment. Subject 13 was definitely the outlier of the group. The clones of each subject were clustered together. This makes sense as they are just different sequences from the same subject. However, it does show that there is a difference between each sequence. In regards to a comparison within the subjects themselves, subject 10 had the closest related clones. However, this subject did have the least clones of the subjects tested. While running a comparison between two subjects, it was found that 4 and 9 were the most closely related out of the others tested. In conclusion, this project allowed us to practice looked at how related sequences are through min and max differences, unrooted trees, and S values.
Defining Research Project
Question
Is there a genetic difference in HIV-1 in patients who contracted the virus sexually or by intravenous injection?
Hypothesis
We hypothesize that there will not be a genetic difference in HIV-1 in patients who contracted the virus sexually or by intravenous injection
Subjects, Visits, Clones
We will use 25 sequences from the moderate progressor category in Markham et al. This will be the best group to compare to outside sequences because they are the most stable group in terms of visits. For the other half of the sequences, we will search more recent studies for sexually contracted HIV-1 sequences. This will be completed during class on 9/26.
Acknowledgments
- I would like to thank my partner, Jordan Detamore, for assistance on this project.
- We collaborated in class on the Activity 2 process together.
- We also worked on our idea for the HIV project together in class.
- I would also like to thank Kam D. Dahlquist, Ph.D. for providing the instructions and information for this assignment both in class and on this document: BIOL368/F16:Week 4.
- Even though I worked with the people noted above, this individual journal entry was completed by me and not copied from another source.
- Matthew K. Oki 18:44, 26 September 2016 (EDT):
References
- Biology Workbench
- BIOL368/F16:Week 4
- Markham, R.B., Wang, W.C., Weisstein, A.E., Wang, Z., Munoz, A., Templeton, A., Margolick, J., Vlahov, D., Quinn, T., Farzadegan, H., & Yu, X.F. (1998). Patterns of HIV-1 evolution in individuals with differing rates of CD4 T cell decline. Proc Natl Acad Sci U S A. 95, 12568-12573. doi: 10.1073/pnas.95.21.12568
- Exploring HIV Evolution
Class Homework Links
Weekly Assignments
Individual Journals
Class Journals
Other Links
My Home Page: Matthew K. Oki
Class Home Page: Bioinformatics Home Page
My Paper Resume: Matthew K. Oki Resume