Matthew K. Oki Individual Journal 11
Week 11 Individual Journal
Purpose
- Our purpose was to continue with our research project, and to explore the amino acid changes in our sequences as compared to Huang et al.
Methods & Results (Project)
- ...(Continued Methods from Week 10)
- A multiple sequences alignment was ran to compare the three most diverse clones to the template from the Huang et al. paper. The alignment is shown in a picture below:
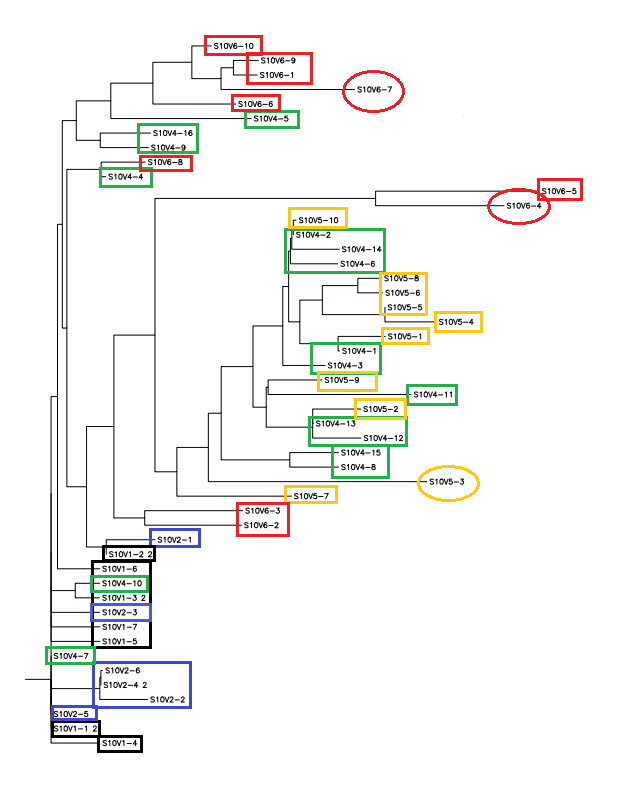
- A phylogenetic tree was postulated through a multiple sequence alignment with all of the clones:
- A 3D model of the "Template" sequence was postulated using the Cn3D program.
- On this 3D model, the "no consensus" changes from the alignment were highlighted. This is shown in the figure below:
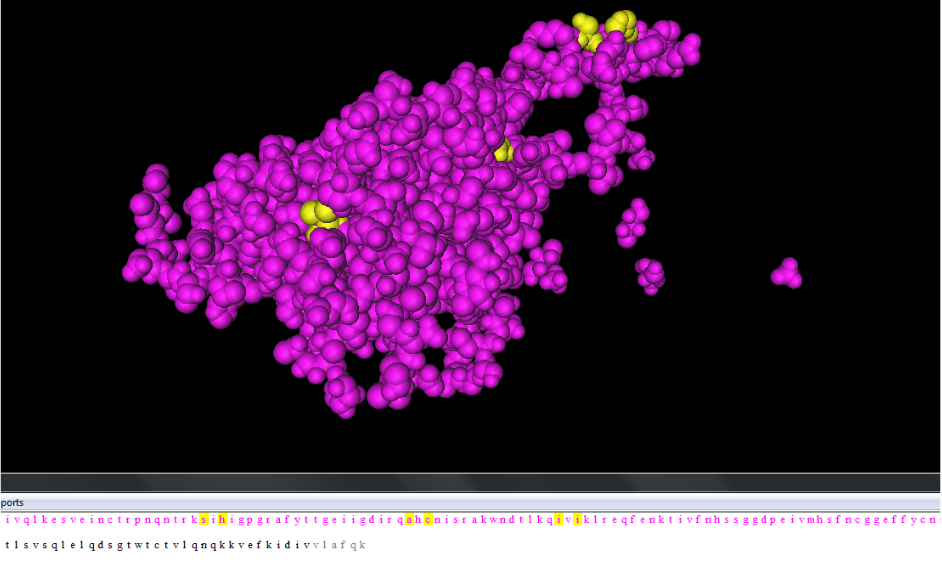
- Two of the changes occurred on the immunogenic tip (shown as the yellow amino acids in the top-right of the photo).
- The left change on the immunogenic tip did not seem to promise any significance, but the change on the right went from a positive histidine to polar uncharged amino acids.
Data and Files
A pdf version of our work-in-progress can be found here.
Scientific Conclusion
In Week 10's methods, S10V6-4, S10V5-3, and S10V6-7 had the highest divergence. Using this data, a multiple sequence alignment was taken to compare these three clones with the template sequence of Huang et al.(2005). This yielded the result that six amino acids had no consensus throughout the different sequences. Among these six, two of them were on the face of the protein and on the immunogenic tip. This is of significance because it is on the ever important immunogenic tip of the virus. One of these amino acid's changes included a change from a positively charged histidine in the template that changed to polar uncharged amino acids in the other sequences. We need to look at amino acids like this more in depth to determine there is indeed significance to the change.
Acknowledgements
- I would like to thank my partner, William P. Fuchs, for the assistance on this week's project, both in assistance on the structure program methods in class and completion of the our project idea outside of class.
- I would also like to thank Kam D. Dahlquist, Ph.D. for providing the instructions and information for this assignment both in class and on this document: BIOL368/F16:Week 11.
- Even though I worked with the people noted above, this individual journal entry was completed by me and not copied from another source.
- Matthew K. Oki 18:07, 14 November 2016 (EST):
References
- Biology Workbench
- BIOL368/F16:Week 11
- ExPASY Translate tool
- UniProt Knowledgebase (UniProt KB)
- PredictProtein server
- Cn3D software site
- Markham, R.B., Wang, W.C., Weisstein, A.E., Wang, Z., Munoz, A., Templeton, A., Margolick, J., Vlahov, D., Quinn, T., Farzadegan, H., & Yu, X.F. (1998). Patterns of HIV-1 evolution in individuals with differing rates of CD4 T cell decline. Proc Natl Acad Sci U S A. 95, 12568-12573. doi: 10.1073/pnas.95.21.12568
- Huang, C. C., Tang, M., Zhang, M. Y., Majeed, S., Montabana, E., Stanfield, R. L., ... & Wyatt, R. (2005). Structure of a V3-containing HIV-1 gp120 core. Science, 310(5750), 1025-1028. doi: 10.1126/science.1118398
Class Homework Links
Weekly Assignments
Individual Journals
Class Journals
Other Links
My Home Page: Matthew K. Oki
Class Home Page: Bioinformatics Home Page
My Paper Resume: Matthew K. Oki Resume