Macie Duran Week 6
Template
User Page
Assignments
- Week 1 Assignment
- Week 2 Assignment
- Week 3 Assignment
- Week 4 Assignment
- Week 5 Assignment
- Week 6 Assignment
- Week 7 Assignment
- Week 8 Assignment
- Week 9 Assignment
- Week 10 Assignment
- Week 11 Assignment
- Week 12 Assignment
- Week 14 Assignment
Journals
- Macie Duran Week 1
- Macie Duran Week 2
- Macie Duran Week 3
- Macie Duran Week 4
- Macie Duran Week 5
- Macie Duran Week 6
- Macie Duran Week 7
- Therapeutic Target Database (TTD) Review
- Macie Duran Week 10
- Macie Duran Week 11
- The Mutants Week 12
- The Mutants Week 14
Class Journals
- Class Journal Week 1
- Class Journal Week 2
- Class Journal Week 3
- Class Journal Week 4
- Class Journal Week 5
- Class Journal Week 6
- Class Journal Week 7
- Class Journal Week 8
- Class Bibliography Week 10
- Class Journal Week 11
- Class Journal Week 14
Purpose
The purpose of this project was to further explore the ACE2 receptor and SARS-CoV-2 spike protein by conducting research. Gaining knowledge on the interaction between ACE2 and SARS-CoV-2 allows us to further understand the viral entry mechanism, and potentially develop preventative measures and treatments for COVID-19.
Methods & Results
Coronavirus Research Project
- I used NCBI GenBank and UniProt to find the ACE2 receptor amino acid sequences for rabbit, cat, ferret, mouse, rat, pig, civet, bat, chimpanzee, human, and opossum (outgroup). The sequences are below.
- Rabbit:
>QHX39726.1 angiotensin I converting enzyme 2 [Oryctolagus cuniculus] MSGSSWLLLSLVAVTAAQSTIEELAKTFLEKFNQEAEDLSYQSALASWDYNTNITEENVQKMNDAEAKWS AFYEEQSKLAKTYPSQEVQNLTVKRQLQALQQSGSSALSADKSKQLNTILSTMSTIYSTGKVCNQSNPQE CFLLEPGLDEIMAKSTDYNERLWAWEGWRSVVGKQLRPLYEEYVVLKNEMARANNYEDYGDYWRADYEAE GADGYDYSRSQLIDDVERTFSEIKPLYEQLHAYVRTKLMDAYPSRISPTGCLPAHLLGDMWGRFWTNLYS LTVPFGQKPNIDVTDTMVNQGWDAERIFKEAEKFFVSVGLPSMTHGFWENSMLPESGDGRKVVCHPTAWD LGKRDFRIKMCTKVTMDNFLTAHHEMGHIQYDMAYATQPFLLRNGANEGFHEAVGEIMSLSAATPEHLKS IGLLPYDFHEDNETEINFLLKQALTIVGTLPFTYMLEKWRWMVFKGEIPKEQWMQKWWEMKREIVGVVEP MPHDETYCDPAALFHVANDYSFIRYYTRTIYQFQFQEALCQAAQHEGPLHKCDISNSTEAGQKLLNMLRL GRSEPWTLALENVVGAKNMDVRPLLNYFEPLFTWLKEQNRNSFVGWSTEWTPYADQSIKVRISLKTALGD QAYEWNDSEMYLFRSSVAYAMRKYFSEVKNQTILFGEEDVRVSDLKPRISFNFFVTAPNNVNDIIPRNEV EEAISMSRSRINDIFRLDDNSLEFVGIQPTLEPPYESPVPIWLVVFGVVMGMIVIGIVVLIFTGIKDRRK QKQAKREENPYGFVDMSKGENNSGFQNSDDIQTSF
>sp|Q56H28|ACE2_FELCA Angiotensin-converting enzyme 2 OS=Felis catus OX=9685 GN=ACE2 PE=2 SV=1 MSGSFWLLLSFAALTAAQSTTEELAKTFLEKFNHEAEELSYQSSLASWNYNTNITDENVQ KMNEAGAKWSAFYEEQSKLAKTYPLAEIHNTTVKRQLQALQQSGSSVLSADKSQRLNTIL NAMSTIYSTGKACNPNNPQECLLLEPGLDDIMENSKDYNERLWAWEGWRAEVGKQLRPLY EEYVALKNEMARANNYEDYGDYWRGDYEEEWTDGYNYSRSQLIKDVEHTFTQIKPLYQHL HAYVRAKLMDTYPSRISPTGCLPAHLLGDMWGRFWTNLYPLTVPFGQKPNIDVTDAMVNQ SWDARRIFKEAEKFFVSVGLPNMTQGFWENSMLTEPGDSRKVVCHPTAWDLGKGDFRIKM CTKVTMDDFLTAHHEMGHIQYDMAYAVQPFLLRNGANEGFHEAVGEIMSLSAATPNHLKT IGLLSPGFSEDSETEINFLLKQALTIVGTLPFTYMLEKWRWMVFKGEIPKEQWMQKWWEM KREIVGVVEPVPHDETYCDPASLFHVANDYSFIRYYTRTIYQFQFQEALCRIAKHEGPLH KCDISNSSEAGKKLLQMLTLGKSKPWTLALEHVVGEKKMNVTPLLKYFEPLFTWLKEQNR NSFVGWNTDWRPYADQSIKVRISLKSALGDEAYEWNDNEMYLFRSSVAYAMREYFSKVKN QTIPFVEDNVWVSNLKPRISFNFFVTASKNVSDVIPRSEVEEAIRMSRSRINDAFRLDDN SLEFLGIQPTLSPPYQPPVTIWLIVFGVVMGVVVVGIVLLIVSGIRNRRKNNQARSEENP YASVDLSKGENNPGFQHADDVQTSF
>BAE53380.1 angiotensin I converting enzyme 2 [Mustela putorius furo] MLGSSWLLLSLAALTAAQSTTEDLAKTFLEKFNYEAEELSYQNSLASWNYNTNITDENIQKMNIAGAKWS AFYEEESQHAKTYPLEEIQDPIIKRQLRALQQSGSSVLSADKRERLNTILNAMSTIYSTGKACNPNNPQE CLLLEPGLDDIMENSKDYNERLWAWEGWRSEVGKQLRPLYEEYVALKNEMARANNYEDYGDYWRGDYEEE WADGYSYSRNQLIEDVEHTFTQIKPLYEHLHAYVRAKLMDAYPSRISPTGCLPAHLLGDMWGRFWTNLYP LMVPFRQKPNIDVTDAMVNQSWDARRIFEEAETFFVSVGLPNMTEGFWQNSMLTEPGDNRKVVCHPTAWD LGKRDFRIKMCTKVTMDDFLTAHHEMGHIQYDMAYAEQPFLLRNGANEGFHEAVGEIMSLSAATPNHLKN IGLLPPDFSEDSETDINFLLKQALTIVGTLPFTYMLEKWRWMVFKGEIPKEQWMQKWWEMKRDIVGVVEP LPHDETYCDPAALFHVANDYSFIRYYTRTIYQFQFQEALCQIAKHEGPLYKCDISNSSEAGQKLHEMLSL GRSKPWTFALERVVGAKTMDVRPLLNYFEPLFTWLKEQNRNSFVGWNTDWSPYADQSIKVRISLKSALGE KAYEWNDNEMYFFQSSIAYAMREYFSKVKNQTIPFVGKDVRVSDLKPRISFNFIVTSPENMSDIIPRADV EEAIRKSRGRINDAFRLDDNSLEFLGIQPTLEPPYQPPVTIWLIVFGVVMGVVVVGIFLLIFSGIRNRRK NNQARSEENPYASVDLSKGENNPGFQNVDDVQTSF
>sp|Q8R0I0|ACE2_MOUSE Angiotensin-converting enzyme 2 OS=Mus musculus OX=10090 GN=Ace2 PE=1 SV=1 MSSSSWLLLSLVAVTTAQSLTEENAKTFLNNFNQEAEDLSYQSSLASWNYNTNITEENAQ KMSEAAAKWSAFYEEQSKTAQSFSLQEIQTPIIKRQLQALQQSGSSALSADKNKQLNTIL NTMSTIYSTGKVCNPKNPQECLLLEPGLDEIMATSTDYNSRLWAWEGWRAEVGKQLRPLY EEYVVLKNEMARANNYNDYGDYWRGDYEAEGADGYNYNRNQLIEDVERTFAEIKPLYEHL HAYVRRKLMDTYPSYISPTGCLPAHLLGDMWGRFWTNLYPLTVPFAQKPNIDVTDAMMNQ GWDAERIFQEAEKFFVSVGLPHMTQGFWANSMLTEPADGRKVVCHPTAWDLGHGDFRIKM CTKVTMDNFLTAHHEMGHIQYDMAYARQPFLLRNGANEGFHEAVGEIMSLSAATPKHLKS IGLLPSDFQEDSETEINFLLKQALTIVGTLPFTYMLEKWRWMVFRGEIPKEQWMKKWWEM KREIVGVVEPLPHDETYCDPASLFHVSNDYSFIRYYTRTIYQFQFQEALCQAAKYNGSLH KCDISNSTEAGQKLLKMLSLGNSEPWTKALENVVGARNMDVKPLLNYFQPLFDWLKEQNR NSFVGWNTEWSPYADQSIKVRISLKSALGANAYEWTNNEMFLFRSSVAYAMRKYFSIIKN QTVPFLEEDVRVSDLKPRVSFYFFVTSPQNVSDVIPRSEVEDAIRMSRGRINDVFGLNDN SLEFLGIHPTLEPPYQPPVTIWLIIFGVVMALVVVGIIILIVTGIKGRKKKNETKREENP YDSMDIGKGESNAGFQNSDDAQTSF
- Rat:
>sp|Q5EGZ1|ACE2_RAT Angiotensin-converting enzyme 2 OS=Rattus norvegicus OX=10116 GN=Ace2 PE=1 SV=1 MSSSCWLLLSLVAVATAQSLIEEKAESFLNKFNQEAEDLSYQSSLASWNYNTNITEENAQ KMNEAAAKWSAFYEEQSKIAQNFSLQEIQNATIKRQLKALQQSGSSALSPDKNKQLNTIL NTMSTIYSTGKVCNSMNPQECFLLEPGLDEIMATSTDYNRRLWAWEGWRAEVGKQLRPLY EEYVVLKNEMARANNYEDYGDYWRGDYEAEGVEGYNYNRNQLIEDVENTFKEIKPLYEQL HAYVRTKLMEVYPSYISPTGCLPAHLLGDMWGRFWTNLYPLTTPFLQKPNIDVTDAMVNQ SWDAERIFKEAEKFFVSVGLPQMTPGFWTNSMLTEPGDDRKVVCHPTAWDLGHGDFRIKM CTKVTMDNFLTAHHEMGHIQYDMAYAKQPFLLRNGANEGFHEAVGEIMSLSAATPKHLKS IGLLPSNFQEDNETEINFLLKQALTIVGTLPFTYMLEKWRWMVFQDKIPREQWTKKWWEM KREIVGVVEPLPHDETYCDPASLFHVSNDYSFIRYYTRTIYQFQFQEALCQAAKHDGPLH KCDISNSTEAGQKLLNMLSLGNSGPWTLALENVVGSRNMDVKPLLNYFQPLFVWLKEQNR NSTVGWSTDWSPYADQSIKVRISLKSALGKNAYEWTDNEMYLFRSSVAYAMREYFSREKN QTVPFGEADVWVSDLKPRVSFNFFVTSPKNVSDIIPRSEVEEAIRMSRGRINDIFGLNDN SLEFLGIYPTLKPPYEPPVTIWLIIFGVVMGTVVVGIVILIVTGIKGRKKKNETKREENP YDSMDIGKGESNAGFQNSDDAQTSF
- Domestic pig (unreviewed UniProt sequence):
>tr|A0A220QT48|A0A220QT48_PIG Angiotensin-converting enzyme OS=Sus scrofa domesticus OX=9825 GN=ACE2 PE=2 SV=1 MSGSFWLLLSLIPVTAAQSTTEELAKTFLEKFNLEAEDLAYQSSLASWNYNTNITDENIQ KMNDARAKWSAFYEEQSRIAKTYPLDEIQTLILKRQLQALQQSGTSGLSADKSKRLNTIL NTMSTIYSSGKVLDPNNPQECLVLEPGLDEIMENSKDYSRRLWAWESWRAEVGKQLRPLY EEYVVLENEMARANNYEDYGDYWRGDYEVTGTGDYDYSRNQLMEDVERTFAEIKPLYEHL HAYVRAKLMDAYPSRISPTGCLPAHLLGDMWGRFWTNLYPLTVPFGEKPSIDVTEAMVNQ SWDAIRIFEEAEKFFVSIGLPNMTQGFWNNSMLTEPGDGRKVVCHPTAWDLGKGDFRIKM CTKVTMDDFLTAHHEMGHIQYDMAYAIQPYLLRNGANEGFHEAVGEIMSLSAATPHYLKA LGLLPPDFYEDSETEINFLLKQALTIVGTLPFTYMLEKWRWMVFKGEIPKEQWMQKWWEM KREIVGVVEPLPHDETYCDPACLFHVAEDYSFIRYYTRTIYQFQFHEALCRTAKHEGPLY KCDISNSTEAGQKLLQMLSLGKSEPWTLALENIVGVKTMDVKPLLSYFEPLLTWLKAQNG NSSVGWNTDWTPYADQSIKVRISLKSALGKEAYEWNDNEMYLFRSSIAYAMRNYFSSAKN ETIPFGAEDVWVSDLKPRISFNFFVTSPANMSDIIPRSDVEKAISMSRSRINDAFRLDDN TLEFLGIQPTLGPPDEPPVTVWLIIFGVVMGLVVVGIVVLIFTGIRDRRKKKQASSEENP YGSMDLSKGESNSGFQNGDDIQTSF
>sp|Q56NL1|ACE2_PAGLA Angiotensin-converting enzyme 2 OS=Paguma larvata OX=9675 GN=ACE2 PE=1 SV=1 MSGSFWLLLSFAALTAAQSTTEELAKTFLETFNYEAQELSYQSSVASWNYNTNITDENAK NMNEAGAKWSAYYEEQSKLAQTYPLAEIQDAKIKRQLQALQQSGSSVLSADKSQRLNTIL NAMSTIYSTGKACNPNNPQECLLLEPGLDNIMENSKDYNERLWAWEGWRAEVGKQLRPLY EEYVALKNEMARANNYEDYGDYWRGDYEEEWTGGYNYSRNQLIQDVEDTFEQIKPLYQHL HAYVRAKLMDTYPSRISRTGCLPAHLLGDMWGRFWTNLYPLTVPFGQKPNIDVTDAMVNQ NWDARRIFKEAEKFFVSVGLPNMTQGFWENSMLTEPGDGRKVVCHPTAWDLGKGDFRIKM CTKVTMDDFLTAHHEMGHIQYDMAYAAQPFLLRNGANEGFHEAVGEIMSLSAATPNHLKT IGLLSPAFSEDNETEINFLLKQALTIVGTLPFTYMLEKWRWMVFKGAIPKEQWMQKWWEM KRNIVGVVEPVPHDETYCDPASLFHVANDYSFIRYYTRTIYQFQFQEALCQIAKHEGPLH KCDISNSTEAGKKLLEMLSLGRSEPWTLALERVVGAKNMNVTPLLNYFEPLFTWLKEQNR NSFVGWDTDWRPYSDQSIKVRISLKSALGEKAYEWNDNEMYLFRSSIAYAMREYFSKVKN QTIPFVEDNVWVSDLKPRISFNFFVTFSNNVSDVIPRSEVEDAIRMSRSRINDAFRLDDN SLEFLGIEPTLSPPYRPPVTIWLIVFGVVMGAIVVGIVLLIVSGIRNRRKNDQAGSEENP YASVDLNKGENNPGFQHADDVQTSF
- Greater rufous horseshoe bat (unreviewed UniProt sequence):
>tr|E2DHI2|E2DHI2_RHIFE Angiotensin-converting enzyme OS=Rhinolophus ferrumequinum OX=59479 GN=ACE2 PE=2 SV=1 MSGSSWFLLSLVAVTAAQSTTEDLAKKFLDDFNSEAENLSHQSSLASWEYNTNISDENVQ KMDEAGAKWSDFYEKQSKLAKNFSLEEIHNDTVKLQLQILQQSGSPVLSEDKSKRLNSIL NAMSTIYSTGKVCKPNNPQECLLLEPGLDNIMGTSKDYNERLWAWEGWRAEVGKQLRPLY EEYVVLKNEMARGYHYEDYGDYWRRDYETEGSPDLEYSRDQLIKDVERIFAEIKPLYEQL HAYVRTKLMDTYPFHISPTGCLPAHLLGDMWGRFWTNLYPLTVPFGQKPNIDVTDAMLNQ NWDAKRIFKEAEKFLVSIGLPNMTEGFWNNSMLTDPGDGRKVVCHPTAWDLGKGDFRIKM CTKVTMEDFLTAHHEMGHIQYDMAYASQPYLLRNGANEGFHEAVGEVMSLSVATPEHLKT MGLLSSDFLEDNETEINFLFKQALNIVGTLPLTYMLEKWRWMVFKGEIPKEEWMKKWWEM KRKIVGVVEPVPHDETYCDPASLFHVANDYSFIRYYTRTIFEFQFHEALCRIAKHDGPLH KCGISNSTDAGEKLHQMLSVGKSQPWTSVLKDFVGSKNMDVGPLLRYFEPLYTWLTEQNR KSFVGWNTDWSPYADQSIKVWISLKSALGEKAYEWNNNEMYLFRSSVAYAMREYFLKTKN QTILFGEEDVWVSNLKPRISFNFYVTSPRNLSDIIPRPEVEGAIRMSRSRINDAFRLDDN SLEFLGIQPTLGPPYQPPVTIWLIVFGVVMAVVVVGIVVLIITGIRDRRKKDQARSEENP YSSVDLSKGENNPGFQNGNDVQTSF
- Chimpanzee (unreviewed UniProt sequence):
>tr|A0A2J8KU96|A0A2J8KU96_PANTR Angiotensin-converting enzyme OS=Pan troglodytes OX=9598 GN=ACE2 PE=3 SV=1 MSGSSWLLLSLVAVTAAQSTIEEQAKTFLDKFNHEAEDLFYQSSLASWNYNTNITEENVQ NMNNAGDKWSAFLKEQSTLAQMYPLQEIQNLTVKLQLQALQQNGSSVLSEDKSKRLNTIL NTMSAIYSTGKVCNPNNPQECLLLEPGLNEIMANSLDYNERLWAWESWRSEVGKQLRPLY EEYVVLKNEMARANHYEDYGDYWRGDYEVNGVDGYDYSRGQLIEDVEHTFEEIKPLYEHL HAYVRAKLMNAYPSYISPIGCLPAHLLGDMWGRFWTNLYSLTVPFGQKPNIDVTDAMVDQ AWDAQRIFKEAEKFFVSVGLPNMTQGFWENSMLTDPGNVQKAVCHPTAWDLGKGDFRILM CTKVTMDDFLTAHHEMGHIQYDMAYAAQPFLLRNGANEGFHEAVGEIMSLSAATPKHLKS IGLLSPDFQEDNETEINFLLKQALTIVGTLPFTYMLEKWRWMVFKGEIPEDQWMKKWWEM KREIVGVVEPVPHDETYCDPASLFHVSNDYSFIRYYTRTLYQFQFQEALCQAAKHEGPLH KCDISNSTEAGQKLFNMLRLGKSEPWTLALENVVGAKNMNVRPLLNYFEPLFTWLKDQNK NSFVGWSTDWSPYADQSIKVRISLKSALGDKAYEWNDNEMYLFRSSVAYAMRQYFLKVKN QMILFGEEDVRVANLKPRISFNFFVTAPKNVSDIIPRTEVEKAIRKSRSRINDAFRLNDN SLEFLGIQPTLGPPNQPPVSIWLIVFGVVMGVIVVGIVILIFTGIRDRKKKNKARSEENP YASVDTSKGENNPGFQNTDDVQTSF
>sp|Q9BYF1|ACE2_HUMAN Angiotensin-converting enzyme 2 OS=Homo sapiens OX=9606 GN=ACE2 PE=1 SV=2 MSSSSWLLLSLVAVTAAQSTIEEQAKTFLDKFNHEAEDLFYQSSLASWNYNTNITEENVQ NMNNAGDKWSAFLKEQSTLAQMYPLQEIQNLTVKLQLQALQQNGSSVLSEDKSKRLNTIL NTMSTIYSTGKVCNPDNPQECLLLEPGLNEIMANSLDYNERLWAWESWRSEVGKQLRPLY EEYVVLKNEMARANHYEDYGDYWRGDYEVNGVDGYDYSRGQLIEDVEHTFEEIKPLYEHL HAYVRAKLMNAYPSYISPIGCLPAHLLGDMWGRFWTNLYSLTVPFGQKPNIDVTDAMVDQ AWDAQRIFKEAEKFFVSVGLPNMTQGFWENSMLTDPGNVQKAVCHPTAWDLGKGDFRILM CTKVTMDDFLTAHHEMGHIQYDMAYAAQPFLLRNGANEGFHEAVGEIMSLSAATPKHLKS IGLLSPDFQEDNETEINFLLKQALTIVGTLPFTYMLEKWRWMVFKGEIPKDQWMKKWWEM KREIVGVVEPVPHDETYCDPASLFHVSNDYSFIRYYTRTLYQFQFQEALCQAAKHEGPLH KCDISNSTEAGQKLFNMLRLGKSEPWTLALENVVGAKNMNVRPLLNYFEPLFTWLKDQNK NSFVGWSTDWSPYADQSIKVRISLKSALGDKAYEWNDNEMYLFRSSVAYAMRQYFLKVKN QMILFGEEDVRVANLKPRISFNFFVTAPKNVSDIIPRTEVEKAIRMSRSRINDAFRLNDN SLEFLGIQPTLGPPNQPPVSIWLIVFGVVMGVIVVGIVILIFTGIRDRKKKNKARSGENP YASIDISKGENNPGFQNTDDVQTSF
- Opossum (outgroup):
>tr|F6WXR7|F6WXR7_MONDO Angiotensin-converting enzyme OS=Monodelphis domestica OX=13616 GN=ACE2 PE=3 SV=2 MLDPLWLFFSLLAVTAAQNSIEEDAKTFLDDYNAKAEELSHQSALASWEYNTNITNENVE KMNEAAARWSSFYENQSSISRTYPLNEITNATVKLQLKSLQKKEGAVLSTEQSVRLNTIL NTMSTLYSTGSVCNSETPQQCFLLEPGLDKIMDESTDYDERLWAWEGWRSKVGKEMRPLY EEYVELKNELAKGNNYEDYGDYWRGDYEVEEPSEYVYSRPQLKKDVENTFKQIKSLYEHL HAYVRRKMRNTYGSLISETGGLPAHLLGDMWGRFWTNLYSLTMPYREKPNIDVTSAMKKQ NWSARRIFQEAEMFFASVGLPNMTEGFWKNSMLTEPNDGRKVVCHPTAWDLGKNDFRIKM CTKVTMDDFLTAHHEMGHIQYDMAYAKQPFTLRNGANEGFHEAVGEIMSLSAATPKHLQA LGLLPPTFQEDNETEINFLFKQALTIIGTMPFTYMLENWRWMVFEGKIPKEEWMKKWWEM KREIVGVVEPLPHDETYCDPAALFHVANDYSFIRYYTRTIYQFQFHKALCKIAQPSAALH KCDITNSTEAGTKLQNMLKMGKSEPWTKALESIVGNKMMDAGPLLEYFEPLFTWLKEQNK DAYVGWNTDWSPYNAYKIKVRISLKTLGENAYTWNENEMYLFQSSIVFAMRQYFLIKKKQ SIPFSNENVKMFDLKPRISFYFFVTFPPNGTSFVPREEVEAAISMSRDRINDAFRLNDNS LEFVGISPTLAPPYEPPVTVWMIVFGVVMGIVVIGIVYLIYTGVRDRKKRAKTSSSNDEN PYVDVDVAGGQHNPAFQSSEDAQTSF
- Then I used Phylogeny.fr to create a multiple sequence alignment.
- I scrolled to the section titled "Phylogeny Analysis" and selected "One Click."
- I pasted the sequences above in FASTA format into the text box and clicked "Submit".
- Of the numbered tabs at the top of the page, I selected 3. Alignment.
- Under the section labeled Output, I selected Alignment in Clustal Format. The alignments are displayed in a text format with symbols indicating conservation between the sequences. "*" indicates invariant conservation, ":" indicates high conservation, "." indicates weak conservation, and a space indicates no conservation.
- I have included the text alignment in clustal format below:
tr|F6WXR7| MLDPLWLFFSLLAVTAAQNSIEEDAKTFLDDYNAKAEELSHQSALASWEYNTNITNENVE
tr|E2DHI2| MSGSSWFLLSLVAVTAAQSTTEDLAKKFLDDFNSEAENLSHQSSLASWEYNTNISDENVQ
sp|Q8R0I0| MSSSSWLLLSLVAVTTAQSLTEENAKTFLNNFNQEAEDLSYQSSLASWNYNTNITEENAQ
sp|Q5EGZ1| MSSSCWLLLSLVAVATAQSLIEEKAESFLNKFNQEAEDLSYQSSLASWNYNTNITEENAQ
tr|A0A220Q MSGSFWLLLSLIPVTAAQSTTEELAKTFLEKFNLEAEDLAYQSSLASWNYNTNITDENIQ
QHX39726.1 MSGSSWLLLSLVAVTAAQSTIEELAKTFLEKFNQEAEDLSYQSALASWDYNTNITEENVQ
tr|A0A2J8K MSGSSWLLLSLVAVTAAQSTIEEQAKTFLDKFNHEAEDLFYQSSLASWNYNTNITEENVQ
sp|Q9BYF1| MSSSSWLLLSLVAVTAAQSTIEEQAKTFLDKFNHEAEDLFYQSSLASWNYNTNITEENVQ
BAE53380.1 MLGSSWLLLSLAALTAAQSTTEDLAKTFLEKFNYEAEELSYQNSLASWNYNTNITDENIQ
sp|Q56NL1| MSGSFWLLLSFAALTAAQSTTEELAKTFLETFNYEAQELSYQSSVASWNYNTNITDENAK
sp|Q56H28| MSGSFWLLLSFAALTAAQSTTEELAKTFLEKFNHEAEELSYQSSLASWNYNTNITDENVQ
* .. *:::*: .:::**. *: *:.**: :* :*::* :*.::***:*****::** :
tr|F6WXR7| KMNEAAARWSSFYENQSSISRTYPLNEITNATVKLQLKSLQKKEGAVLSTEQSVRLNTIL
tr|E2DHI2| KMDEAGAKWSDFYEKQSKLAKNFSLEEIHNDTVKLQLQILQQSGSPVLSEDKSKRLNSIL
sp|Q8R0I0| KMSEAAAKWSAFYEEQSKTAQSFSLQEIQTPIIKRQLQALQQSGSSALSADKNKQLNTIL
sp|Q5EGZ1| KMNEAAAKWSAFYEEQSKIAQNFSLQEIQNATIKRQLKALQQSGSSALSPDKNKQLNTIL
tr|A0A220Q KMNDARAKWSAFYEEQSRIAKTYPLDEIQTLILKRQLQALQQSGTSGLSADKSKRLNTIL
QHX39726.1 KMNDAEAKWSAFYEEQSKLAKTYPSQEVQNLTVKRQLQALQQSGSSALSADKSKQLNTIL
tr|A0A2J8K NMNNAGDKWSAFLKEQSTLAQMYPLQEIQNLTVKLQLQALQQNGSSVLSEDKSKRLNTIL
sp|Q9BYF1| NMNNAGDKWSAFLKEQSTLAQMYPLQEIQNLTVKLQLQALQQNGSSVLSEDKSKRLNTIL
BAE53380.1 KMNIAGAKWSAFYEEESQHAKTYPLEEIQDPIIKRQLRALQQSGSSVLSADKRERLNTIL
sp|Q56NL1| NMNEAGAKWSAYYEEQSKLAQTYPLAEIQDAKIKRQLQALQQSGSSVLSADKSQRLNTIL
sp|Q56H28| KMNEAGAKWSAFYEEQSKLAKTYPLAEIHNTTVKRQLQALQQSGSSVLSADKSQRLNTIL
:*. * .** : :::* :. :. *: :* **. **:. . ** :: .**:**
tr|F6WXR7| NTMSTLYSTGSVCNSETPQQCFLLEPGLDKIMDESTDYDERLWAWEGWRSKVGKEMRPLY
tr|E2DHI2| NAMSTIYSTGKVCKPNNPQECLLLEPGLDNIMGTSKDYNERLWAWEGWRAEVGKQLRPLY
sp|Q8R0I0| NTMSTIYSTGKVCNPKNPQECLLLEPGLDEIMATSTDYNSRLWAWEGWRAEVGKQLRPLY
sp|Q5EGZ1| NTMSTIYSTGKVCNSMNPQECFLLEPGLDEIMATSTDYNRRLWAWEGWRAEVGKQLRPLY
tr|A0A220Q NTMSTIYSSGKVLDPNNPQECLVLEPGLDEIMENSKDYSRRLWAWESWRAEVGKQLRPLY
QHX39726.1 STMSTIYSTGKVCNQSNPQECFLLEPGLDEIMAKSTDYNERLWAWEGWRSVVGKQLRPLY
tr|A0A2J8K NTMSAIYSTGKVCNPNNPQECLLLEPGLNEIMANSLDYNERLWAWESWRSEVGKQLRPLY
sp|Q9BYF1| NTMSTIYSTGKVCNPDNPQECLLLEPGLNEIMANSLDYNERLWAWESWRSEVGKQLRPLY
BAE53380.1 NAMSTIYSTGKACNPNNPQECLLLEPGLDDIMENSKDYNERLWAWEGWRSEVGKQLRPLY
sp|Q56NL1| NAMSTIYSTGKACNPNNPQECLLLEPGLDNIMENSKDYNERLWAWEGWRAEVGKQLRPLY
sp|Q56H28| NAMSTIYSTGKACNPNNPQECLLLEPGLDDIMENSKDYNERLWAWEGWRAEVGKQLRPLY
.:**::**:*.. . .**:*::*****:.** * **. ******.**: ***::****
tr|F6WXR7| EEYVELKNELAKGNNYEDYGDYWRGDYEVEEPSEYVYSRPQLKKDVENTFKQIKSLYEHL
tr|E2DHI2| EEYVVLKNEMARGYHYEDYGDYWRRDYETEGSPDLEYSRDQLIKDVERIFAEIKPLYEQL
sp|Q8R0I0| EEYVVLKNEMARANNYNDYGDYWRGDYEAEGADGYNYNRNQLIEDVERTFAEIKPLYEHL
sp|Q5EGZ1| EEYVVLKNEMARANNYEDYGDYWRGDYEAEGVEGYNYNRNQLIEDVENTFKEIKPLYEQL
tr|A0A220Q EEYVVLENEMARANNYEDYGDYWRGDYEVTGTGDYDYSRNQLMEDVERTFAEIKPLYEHL
QHX39726.1 EEYVVLKNEMARANNYEDYGDYWRADYEAEGADGYDYSRSQLIDDVERTFSEIKPLYEQL
tr|A0A2J8K EEYVVLKNEMARANHYEDYGDYWRGDYEVNGVDGYDYSRGQLIEDVEHTFEEIKPLYEHL
sp|Q9BYF1| EEYVVLKNEMARANHYEDYGDYWRGDYEVNGVDGYDYSRGQLIEDVEHTFEEIKPLYEHL
BAE53380.1 EEYVALKNEMARANNYEDYGDYWRGDYEEEWADGYSYSRNQLIEDVEHTFTQIKPLYEHL
sp|Q56NL1| EEYVALKNEMARANNYEDYGDYWRGDYEEEWTGGYNYSRNQLIQDVEDTFEQIKPLYQHL
sp|Q56H28| EEYVALKNEMARANNYEDYGDYWRGDYEEEWTDGYNYSRSQLIKDVEHTFTQIKPLYQHL
**** *:**:*.. :*:******* *** *.* ** .*** * :**.**::*
tr|F6WXR7| HAYVRRKMRNTYGSLISETGGLPAHLLGDMWGRFWTNLYSLTMPYREKPNIDVTSAMKKQ
tr|E2DHI2| HAYVRTKLMDTYPFHISPTGCLPAHLLGDMWGRFWTNLYPLTVPFGQKPNIDVTDAMLNQ
sp|Q8R0I0| HAYVRRKLMDTYPSYISPTGCLPAHLLGDMWGRFWTNLYPLTVPFAQKPNIDVTDAMMNQ
sp|Q5EGZ1| HAYVRTKLMEVYPSYISPTGCLPAHLLGDMWGRFWTNLYPLTTPFLQKPNIDVTDAMVNQ
tr|A0A220Q HAYVRAKLMDAYPSRISPTGCLPAHLLGDMWGRFWTNLYPLTVPFGEKPSIDVTEAMVNQ
QHX39726.1 HAYVRTKLMDAYPSRISPTGCLPAHLLGDMWGRFWTNLYSLTVPFGQKPNIDVTDTMVNQ
tr|A0A2J8K HAYVRAKLMNAYPSYISPIGCLPAHLLGDMWGRFWTNLYSLTVPFGQKPNIDVTDAMVDQ
sp|Q9BYF1| HAYVRAKLMNAYPSYISPIGCLPAHLLGDMWGRFWTNLYSLTVPFGQKPNIDVTDAMVDQ
BAE53380.1 HAYVRAKLMDAYPSRISPTGCLPAHLLGDMWGRFWTNLYPLMVPFRQKPNIDVTDAMVNQ
sp|Q56NL1| HAYVRAKLMDTYPSRISRTGCLPAHLLGDMWGRFWTNLYPLTVPFGQKPNIDVTDAMVNQ
sp|Q56H28| HAYVRAKLMDTYPSRISPTGCLPAHLLGDMWGRFWTNLYPLTVPFGQKPNIDVTDAMVNQ
***** *: :.* ** * ******************.* *: :**.****.:* .*
tr|F6WXR7| NWSARRIFQEAEMFFASVGLPNMTEGFWKNSMLTEPNDGRKVVCHPTAWDLGKNDFRIKM
tr|E2DHI2| NWDAKRIFKEAEKFLVSIGLPNMTEGFWNNSMLTDPGDGRKVVCHPTAWDLGKGDFRIKM
sp|Q8R0I0| GWDAERIFQEAEKFFVSVGLPHMTQGFWANSMLTEPADGRKVVCHPTAWDLGHGDFRIKM
sp|Q5EGZ1| SWDAERIFKEAEKFFVSVGLPQMTPGFWTNSMLTEPGDDRKVVCHPTAWDLGHGDFRIKM
tr|A0A220Q SWDAIRIFEEAEKFFVSIGLPNMTQGFWNNSMLTEPGDGRKVVCHPTAWDLGKGDFRIKM
QHX39726.1 GWDAERIFKEAEKFFVSVGLPSMTHGFWENSMLPESGDGRKVVCHPTAWDLGKRDFRIKM
tr|A0A2J8K AWDAQRIFKEAEKFFVSVGLPNMTQGFWENSMLTDPGNVQKAVCHPTAWDLGKGDFRILM
sp|Q9BYF1| AWDAQRIFKEAEKFFVSVGLPNMTQGFWENSMLTDPGNVQKAVCHPTAWDLGKGDFRILM
BAE53380.1 SWDARRIFEEAETFFVSVGLPNMTEGFWQNSMLTEPGDNRKVVCHPTAWDLGKRDFRIKM
sp|Q56NL1| NWDARRIFKEAEKFFVSVGLPNMTQGFWENSMLTEPGDGRKVVCHPTAWDLGKGDFRIKM
sp|Q56H28| SWDARRIFKEAEKFFVSVGLPNMTQGFWENSMLTEPGDSRKVVCHPTAWDLGKGDFRIKM
*.* ***:*** *:.*:*** ** *** ****.:. : .*.**********: **** *
tr|F6WXR7| CTKVTMDDFLTAHHEMGHIQYDMAYAKQPFTLRNGANEGFHEAVGEIMSLSAATPKHLQA
tr|E2DHI2| CTKVTMEDFLTAHHEMGHIQYDMAYASQPYLLRNGANEGFHEAVGEVMSLSVATPEHLKT
sp|Q8R0I0| CTKVTMDNFLTAHHEMGHIQYDMAYARQPFLLRNGANEGFHEAVGEIMSLSAATPKHLKS
sp|Q5EGZ1| CTKVTMDNFLTAHHEMGHIQYDMAYAKQPFLLRNGANEGFHEAVGEIMSLSAATPKHLKS
tr|A0A220Q CTKVTMDDFLTAHHEMGHIQYDMAYAIQPYLLRNGANEGFHEAVGEIMSLSAATPHYLKA
QHX39726.1 CTKVTMDNFLTAHHEMGHIQYDMAYATQPFLLRNGANEGFHEAVGEIMSLSAATPEHLKS
tr|A0A2J8K CTKVTMDDFLTAHHEMGHIQYDMAYAAQPFLLRNGANEGFHEAVGEIMSLSAATPKHLKS
sp|Q9BYF1| CTKVTMDDFLTAHHEMGHIQYDMAYAAQPFLLRNGANEGFHEAVGEIMSLSAATPKHLKS
BAE53380.1 CTKVTMDDFLTAHHEMGHIQYDMAYAEQPFLLRNGANEGFHEAVGEIMSLSAATPNHLKN
sp|Q56NL1| CTKVTMDDFLTAHHEMGHIQYDMAYAAQPFLLRNGANEGFHEAVGEIMSLSAATPNHLKT
sp|Q56H28| CTKVTMDDFLTAHHEMGHIQYDMAYAVQPFLLRNGANEGFHEAVGEIMSLSAATPNHLKT
******::****************** **: ***************:****.*** :*:
tr|F6WXR7| LGLLPPTFQEDNETEINFLFKQALTIIGTMPFTYMLENWRWMVFEGKIPKEEWMKKWWEM
tr|E2DHI2| MGLLSSDFLEDNETEINFLFKQALNIVGTLPLTYMLEKWRWMVFKGEIPKEEWMKKWWEM
sp|Q8R0I0| IGLLPSDFQEDSETEINFLLKQALTIVGTLPFTYMLEKWRWMVFRGEIPKEQWMKKWWEM
sp|Q5EGZ1| IGLLPSNFQEDNETEINFLLKQALTIVGTLPFTYMLEKWRWMVFQDKIPREQWTKKWWEM
tr|A0A220Q LGLLPPDFYEDSETEINFLLKQALTIVGTLPFTYMLEKWRWMVFKGEIPKEQWMQKWWEM
QHX39726.1 IGLLPYDFHEDNETEINFLLKQALTIVGTLPFTYMLEKWRWMVFKGEIPKEQWMQKWWEM
tr|A0A2J8K IGLLSPDFQEDNETEINFLLKQALTIVGTLPFTYMLEKWRWMVFKGEIPEDQWMKKWWEM
sp|Q9BYF1| IGLLSPDFQEDNETEINFLLKQALTIVGTLPFTYMLEKWRWMVFKGEIPKDQWMKKWWEM
BAE53380.1 IGLLPPDFSEDSETDINFLLKQALTIVGTLPFTYMLEKWRWMVFKGEIPKEQWMQKWWEM
sp|Q56NL1| IGLLSPAFSEDNETEINFLLKQALTIVGTLPFTYMLEKWRWMVFKGAIPKEQWMQKWWEM
sp|Q56H28| IGLLSPGFSEDSETEINFLLKQALTIVGTLPFTYMLEKWRWMVFKGEIPKEQWMQKWWEM
:***. * **.**:****:****.*:**:*:*****:****** . ** ::* :*****
tr|F6WXR7| KREIVGVVEPLPHDETYCDPAALFHVANDYSFIRYYTRTIYQFQFHKALCKIAQPSAALH
tr|E2DHI2| KRKIVGVVEPVPHDETYCDPASLFHVANDYSFIRYYTRTIFEFQFHEALCRIAKHDGPLH
sp|Q8R0I0| KREIVGVVEPLPHDETYCDPASLFHVSNDYSFIRYYTRTIYQFQFQEALCQAAKYNGSLH
sp|Q5EGZ1| KREIVGVVEPLPHDETYCDPASLFHVSNDYSFIRYYTRTIYQFQFQEALCQAAKHDGPLH
tr|A0A220Q KREIVGVVEPLPHDETYCDPACLFHVAEDYSFIRYYTRTIYQFQFHEALCRTAKHEGPLY
QHX39726.1 KREIVGVVEPMPHDETYCDPAALFHVANDYSFIRYYTRTIYQFQFQEALCQAAQHEGPLH
tr|A0A2J8K KREIVGVVEPVPHDETYCDPASLFHVSNDYSFIRYYTRTLYQFQFQEALCQAAKHEGPLH
sp|Q9BYF1| KREIVGVVEPVPHDETYCDPASLFHVSNDYSFIRYYTRTLYQFQFQEALCQAAKHEGPLH
BAE53380.1 KRDIVGVVEPLPHDETYCDPAALFHVANDYSFIRYYTRTIYQFQFQEALCQIAKHEGPLY
sp|Q56NL1| KRNIVGVVEPVPHDETYCDPASLFHVANDYSFIRYYTRTIYQFQFQEALCQIAKHEGPLH
sp|Q56H28| KREIVGVVEPVPHDETYCDPASLFHVANDYSFIRYYTRTIYQFQFQEALCRIAKHEGPLH
**.*******:**********.****::***********:::***::***. *: ...*:
tr|F6WXR7| KCDITNSTEAGTKLQNMLKMGKSEPWTKALESIVGNKMMDAGPLLEYFEPLFTWLKEQNK
tr|E2DHI2| KCGISNSTDAGEKLHQMLSVGKSQPWTSVLKDFVGSKNMDVGPLLRYFEPLYTWLTEQNR
sp|Q8R0I0| KCDISNSTEAGQKLLKMLSLGNSEPWTKALENVVGARNMDVKPLLNYFQPLFDWLKEQNR
sp|Q5EGZ1| KCDISNSTEAGQKLLNMLSLGNSGPWTLALENVVGSRNMDVKPLLNYFQPLFVWLKEQNR
tr|A0A220Q KCDISNSTEAGQKLLQMLSLGKSEPWTLALENIVGVKTMDVKPLLSYFEPLLTWLKAQNG
QHX39726.1 KCDISNSTEAGQKLLNMLRLGRSEPWTLALENVVGAKNMDVRPLLNYFEPLFTWLKEQNR
tr|A0A2J8K KCDISNSTEAGQKLFNMLRLGKSEPWTLALENVVGAKNMNVRPLLNYFEPLFTWLKDQNK
sp|Q9BYF1| KCDISNSTEAGQKLFNMLRLGKSEPWTLALENVVGAKNMNVRPLLNYFEPLFTWLKDQNK
BAE53380.1 KCDISNSSEAGQKLHEMLSLGRSKPWTFALERVVGAKTMDVRPLLNYFEPLFTWLKEQNR
sp|Q56NL1| KCDISNSTEAGKKLLEMLSLGRSEPWTLALERVVGAKNMNVTPLLNYFEPLFTWLKEQNR
sp|Q56H28| KCDISNSSEAGKKLLQMLTLGKSKPWTLALEHVVGEKKMNVTPLLKYFEPLFTWLKEQNR
**.*:**::** ** :** :*.* *** .*: .** . *:. *** **:** **. **
tr|F6WXR7| DAYVGWNTDWSPYNAYKIKVRISLKT-LGENAYTWNENEMYLFQSSIVFAMRQYFLIKKK
tr|E2DHI2| KSFVGWNTDWSPYADQSIKVWISLKSALGEKAYEWNNNEMYLFRSSVAYAMREYFLKTKN
sp|Q8R0I0| NSFVGWNTEWSPYADQSIKVRISLKSALGANAYEWTNNEMFLFRSSVAYAMRKYFSIIKN
sp|Q5EGZ1| NSTVGWSTDWSPYADQSIKVRISLKSALGKNAYEWTDNEMYLFRSSVAYAMREYFSREKN
tr|A0A220Q NSSVGWNTDWTPYADQSIKVRISLKSALGKEAYEWNDNEMYLFRSSIAYAMRNYFSSAKN
QHX39726.1 NSFVGWSTEWTPYADQSIKVRISLKTALGDQAYEWNDSEMYLFRSSVAYAMRKYFSEVKN
tr|A0A2J8K NSFVGWSTDWSPYADQSIKVRISLKSALGDKAYEWNDNEMYLFRSSVAYAMRQYFLKVKN
sp|Q9BYF1| NSFVGWSTDWSPYADQSIKVRISLKSALGDKAYEWNDNEMYLFRSSVAYAMRQYFLKVKN
BAE53380.1 NSFVGWNTDWSPYADQSIKVRISLKSALGEKAYEWNDNEMYFFQSSIAYAMREYFSKVKN
sp|Q56NL1| NSFVGWDTDWRPYSDQSIKVRISLKSALGEKAYEWNDNEMYLFRSSIAYAMREYFSKVKN
sp|Q56H28| NSFVGWNTDWRPYADQSIKVRISLKSALGDEAYEWNDNEMYLFRSSVAYAMREYFSKVKN
.: ***.*:* ** .***.****: ** :** *.:.**::*.**:.:***:** *:
tr|F6WXR7| QSIPFSNENVKMFDLKPRISFYFFVTFPPNGTSFVPREEVEAAISMSRDRINDAFRLNDN
tr|E2DHI2| QTILFGEEDVWVSNLKPRISFNFYVTSPRNLSDIIPRPEVEGAIRMSRSRINDAFRLDDN
sp|Q8R0I0| QTVPFLEEDVRVSDLKPRVSFYFFVTSPQNVSDVIPRSEVEDAIRMSRGRINDVFGLNDN
sp|Q5EGZ1| QTVPFGEADVWVSDLKPRVSFNFFVTSPKNVSDIIPRSEVEEAIRMSRGRINDIFGLNDN
tr|A0A220Q ETIPFGAEDVWVSDLKPRISFNFFVTSPANMSDIIPRSDVEKAISMSRSRINDAFRLDDN
QHX39726.1 QTILFGEEDVRVSDLKPRISFNFFVTAPNNVNDIIPRNEVEEAISMSRSRINDIFRLDDN
tr|A0A2J8K QMILFGEEDVRVANLKPRISFNFFVTAPKNVSDIIPRTEVEKAIRKSRSRINDAFRLNDN
sp|Q9BYF1| QMILFGEEDVRVANLKPRISFNFFVTAPKNVSDIIPRTEVEKAIRMSRSRINDAFRLNDN
BAE53380.1 QTIPFVGKDVRVSDLKPRISFNFIVTSPENMSDIIPRADVEEAIRKSRGRINDAFRLDDN
sp|Q56NL1| QTIPFVEDNVWVSDLKPRISFNFFVTFSNNVSDVIPRSEVEDAIRMSRSRINDAFRLDDN
sp|Q56H28| QTIPFVEDNVWVSNLKPRISFNFFVTASKNVSDVIPRSEVEEAIRMSRSRINDAFRLDDN
: : * :*.: :****:** * ** . * ...:** :** ** **.**** * *:**
tr|F6WXR7| SLEFVGISPTLAPPYEPPVTVWMIVFGVVMGIVVIGIVYLIYTGVRDRKKRAKTSSSNDE
tr|E2DHI2| SLEFLGIQPTLGPPYQPPVTIWLIVFGVVMAVVVVGIVVLIITGIRDRRKKDQARS--EE
sp|Q8R0I0| SLEFLGIHPTLEPPYQPPVTIWLIIFGVVMALVVVGIIILIVTGIKGRKKKNETKR--EE
sp|Q5EGZ1| SLEFLGIYPTLKPPYEPPVTIWLIIFGVVMGTVVVGIVILIVTGIKGRKKKNETKR--EE
tr|A0A220Q TLEFLGIQPTLGPPDEPPVTVWLIIFGVVMGLVVVGIVVLIFTGIRDRRKKKQASS--EE
QHX39726.1 SLEFVGIQPTLEPPYESPVPIWLVVFGVVMGMIVIGIVVLIFTGIKDRRKQKQAKR--EE
tr|A0A2J8K SLEFLGIQPTLGPPNQPPVSIWLIVFGVVMGVIVVGIVILIFTGIRDRKKKNKARS--EE
sp|Q9BYF1| SLEFLGIQPTLGPPNQPPVSIWLIVFGVVMGVIVVGIVILIFTGIRDRKKKNKARS--GE
BAE53380.1 SLEFLGIQPTLEPPYQPPVTIWLIVFGVVMGVVVVGIFLLIFSGIRNRRKNNQARS--EE
sp|Q56NL1| SLEFLGIEPTLSPPYRPPVTIWLIVFGVVMGAIVVGIVLLIVSGIRNRRKNDQAGS--EE
sp|Q56H28| SLEFLGIQPTLSPPYQPPVTIWLIVFGVVMGVVVVGIVLLIVSGIRNRRKNNQARS--EE
:***:** *** ** .**.:*:::*****. :*:**. ** :*:..*.*. :: *
tr|F6WXR7| NPYVDVDVAGGQHNPAFQSSEDAQTSF
tr|E2DHI2| NPYSSVDLSKGENNPGFQNGNDVQTSF
sp|Q8R0I0| NPYDSMDIGKGESNAGFQNSDDAQTSF
sp|Q5EGZ1| NPYDSMDIGKGESNAGFQNSDDAQTSF
tr|A0A220Q NPYGSMDLSKGESNSGFQNGDDIQTSF
QHX39726.1 NPYGFVDMSKGENNSGFQNSDDIQTSF
tr|A0A2J8K NPYASVDTSKGENNPGFQNTDDVQTSF
sp|Q9BYF1| NPYASIDISKGENNPGFQNTDDVQTSF
BAE53380.1 NPYASVDLSKGENNPGFQNVDDVQTSF
sp|Q56NL1| NPYASVDLNKGENNPGFQHADDVQTSF
sp|Q56H28| NPYASVDLSKGENNPGFQHADDVQTSF
*** :* *: *..** :* ****
- Based on the alignment above, the ACE2 sequence is highly conserved among mammals. This is indicated by the great amount of "*" and ":" symbols below the alignment. There are not many spaces or "." symbols in the alignment.
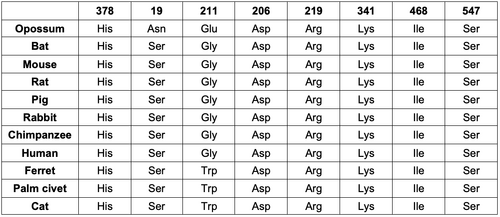
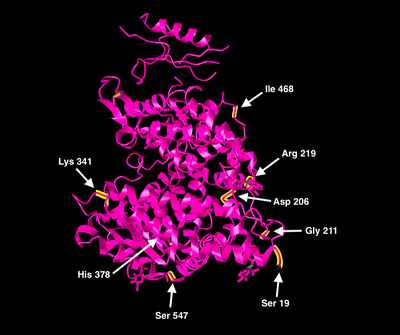
- I decided to analyze the conservation of the amino acids in the key positions 378, 19, 211, 206, 219, 341, 468, and 547. These amino acids were identified as key residues for structure and function of the human ACE2 receptor in a study about genetic variation in human ACE2 that was published in June.
- Six of the amino acids are invariantly conserved in the 11 species.
- One of the amino acids showed weak conservation in the alignment. However, the only species that showed a variance was opossum, which is serving as the outgroup.
- One amino acid showed no conservation between the 11 mammals.
- I also made a table of the 8 key residue positions for the 11 mammals that my partner and I studied.
- Because the majority of the residues were highly conserved, my findings further indicate the importance of these residues to structure and function of ACE2.
- Then, I navigated back to the results page and selected the tab that said 6. Tree Rendering to generate a phylogenetic tree of the sequences.
- I saved the phylogenetic tree and have included it below:
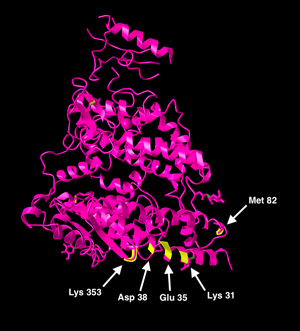
- The key structural and functional residues that we analyzed were distinct from the key residues for viral binding from the Wan et al. article. I used the NCBI Structure Database to create two ACE2 models, one highlighting the viral-binding hotspots, and the other highlighting the eight structural and functional residues. I used the 1R42 model for the figures below.
- Because the structural residues that we analyzed were all different from the viral-binding residues for coronaviruses, for future research, I think it would be interesting to learn more about how ACE2 could potentially be targeted by SARS-CoV-2 treatments.
Presentation Slides
Scientific Conclusion
The eight identified ACE2 structural and functional residues in humans are highly conserved in a variety of mammals, further supporting their importance. The structural and functional residues that were analyzed are distinct from the key viral-binding residues identified in the Wan et al. paper. Knowledge on the ACE2 receptor sequences are helpful for further understanding the receptor's interaction with SARS-CoV-2, and could potentially lead to the development of treatments for COVID19.
Acknowledgments
- I contacted my partner, Nathan R. Beshai, over Zoom and text to discuss our research project.
- I copied and modified the protocol outlined on the Week 6 Assignment page.
- I used the Wan et al. article for background information for my research.
- I used the Guo et al. article for background information for my research.
- I used UniProt Knowledgebase to find entries of ACE2 protein sequences.
- I used NCBI GenBank to find entries of ACE2 protein sequences.
- I used Phylogeny.fr to generate a multiple sequence alignment and phylogenetic tree.
- I used the NCBI iCn3D viewer to visualize and analyze human ACE2 protein structure.
- Except for what is noted above, this individual journal entry was completed by me and not copied from another source.
(Macie Duran (talk) 19:44, 14 October 2020 (PDT))
References
- Angiotensin I converting enzyme 2 [Mustela putorius furo] - Protein - NCBI. (n.d.). Retrieved October 8, 2020, from https://www.ncbi.nlm.nih.gov/protein/BAE53380.1
- Angiotensin I converting enzyme 2 [Oryctolagus cuniculus] - Protein - NCBI. (n.d.). Retrieved October 8, 2020, from https://www.ncbi.nlm.nih.gov/protein/QHX39726.1
- Angiotensin-converting enzyme - A0A220QT48. (2020, October 07). Retrieved October 8, 2020, from https://www.uniprot.org/uniprot/A0A220QT48
- Angiotensin-converting enzyme - A0A2J8KU96. (2020, October 07). Retrieved October 8, 2020, from https://www.uniprot.org/uniprot/A0A2J8KU96
- Angiotensin-converting enzyme - E2DHI2. (2020, October 07). Retrieved October 8, 2020, from https://www.uniprot.org/uniprot/E2DHI2
- Angiotensin-converting enzyme - F6WXR7. (2020, October 07). Retrieved October 8, 2020, from https://www.uniprot.org/uniprot/F6WXR7
- Angiotensin-converting enzyme 2 - Q56H28. (2020, October 07). Retrieved October 8, 2020, from https://www.uniprot.org/uniprot/Q56H28
- Angiotensin-converting enzyme 2 - Q56NL1. (2020, August 12). Retrieved October 8, 2020, from https://www.uniprot.org/uniprot/Q56NL1
- Angiotensin-converting enzyme 2 - Q5EGZ1. (2020, October 07). Retrieved October 8, 2020, from https://www.uniprot.org/uniprot/Q5EGZ1
- Angiotensin-converting enzyme 2 - Q8R0I0. (2020, October 07). Retrieved October 8, 2020, from https://www.uniprot.org/uniprot/Q8R0I0
- Angiotensin-converting enzyme 2 - Q9BYF1. (2020, October 07). Retrieved October 8, 2020, from https://www.uniprot.org/uniprot/Q9BYF1
- Guo, X., Chen, Z., Xia, Y. et al. Investigation of the genetic variation in ACE2 on the structural recognition by the novel coronavirus (SARS-CoV-2). J Transl Med 18, 321 (2020). https://doi.org/10.1186/s12967-020-02486-7
- iCn3D: Web-based 3D Structure Viewer. (n.d.). Retrieved October 14, 2020, from https://www.ncbi.nlm.nih.gov/Structure/icn3d/full.html
- OpenWetWare. (2020). BIOL368/F20:Week 6. Retrieved 14 October 2020, from https://openwetware.org/wiki/BIOL368/F20:Week_6
- Phylogeny.fr: "One Click" Mode. (2020). Retrieved 8 October 2020, from http://www.phylogeny.fr/simple_phylogeny.cgi?workflow_id=b9c0813cbbe9695d63cf7e31da5f026d&tab_index=1
- Wan, Y., Shang, J., Graham, R., Baric, R., & Li, F. (2020). Receptor Recognition by the Novel Coronavirus from Wuhan: an Analysis Based on Decade-Long Structural Studies of SARS Coronavirus. Journal Of Virology, 94(7). doi: 10.1128/jvi.00127-20