Ian R. Wright Week 5
Ian Wright's Bioinformatics Portfolio
Assignment Pages
Individual Journal Entries
- Ian R. Wright Week 1
- Ian R. Wright Week 2
- Ian R. Wright Week 3
- Ian R. Wright Week 4
- Ian R. Wright Week 5
- Ian R. Wright Week 6
- Ian R. Wright Week 7
- Therapeutic Target Database (TTD) Review
- Ian R. Wright Week 9
- Ian R. Wright Week 10
- Ian R. Wright Week 11
- The D614G Research Group Week 12
- Ian R. Wright Week 14
- The D614G Research Group Week 14
Class Journals
BIOL368/F20:Class_Journal_Week_1
BIOL368/F20:Class_Journal_Week_2
BIOL368/F20:Class_Journal_Week_3
BIOL368/F20:Class_Journal_Week_4
BIOL368/F20:Class_Journal_Week_5
BIOL368/F20:Class_Journal_Week_6
BIOL368/F20:Class_Journal_Week_7
BIOL368/F20:Class_Journal_Week_8
BIOL368/F20:Class_Journal_Week_9
BIOL368/F20:Class_Journal_Week_10
BIOL368/F20:Class_Journal_Week_11
BIOL368/F20:Class_Journal_Week_12
BIOL368/F20:Class_Journal_Week_14
Purpose
The NCBI 3D protein structure web application will be explored and used to achieve the goals of this lab. Different styles and color schemes of 3D protein models will highlight certain aspects of the protein. Finally, this knowledge will be used to recreate Figure 4B from Wan et al and attempt to map the interactions between the critical residues in the receptor binding motif of the civet ACE-2 binding spike protein.
Methods and Results
Exploring the Structure of the Spike Protein
- UniProt Knowledgebase (UniProt KB) was used to explore the structure of the SARS-CoV-2 spike protein.
- In the search bar at the top of the UniProt homepage, SARS-CoV-2 was typed and searched.
- The second reviewed protein sequence (marked by a brown page with a star) was listed as ProteinName: 'Spike glycoprotein' and Entry: 'P0DTC2'. This sequence was selected and reviewed.
- The information listed for SARS-CoV-2 spike glycoprotein are as follows:
- Function
- Molecular function
- Biological processes
- Names and Taxonomy
- Subcellular Location
- Pathology and Biotech information
- Post-Translational Modifications (PTM) and Processing
- Interactions
- Structure
- Family and Domains
- Sequence
- Similar Proteins
- Cross-References
- Function
- The information listed for SARS-CoV-2 spike glycoprotein are as follows:
- SARS-CoV RBD optimized for binding to human ACE-2 protein was selected for structure analysis in NCBI protein structure database: Optimized SARS Spike Protein
- Protein was rotated to match the orientation in Figure 1A of Wan et al 2020.
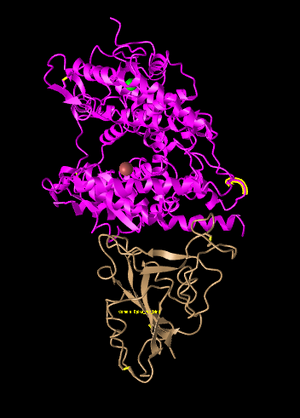
Figure 1: SARS-CoV-2 spike protein (bottom) with human ACE-2 (top). Recreation of Figure 1A from Wan et al 2020.
- Both ACE-2 protein and Spike Protein are shown with their interactions in this database entry.
- Tertiary structure is shown with the locations and interactions of Alpha-helices and Beta-pleated sheets.
- To identify number of domains and their respective locations within ACE-2, the UniProt entry for Human ACE-2 was consulted.
- Extracellular ACE-2 consists of 2 domains, the first of which is a Zinc-metallopeptidase. This domain consists of residues 19-611.
- The NCBI structure database being used only includes residues 1-603 so ACE-2 only has one domain present.
- UniProt entry for SARS2 Spike Protein says that the receptor binding domain is residues 334-527 (194 residues).
- The interacting SARS optimized spike protein in the NCBI structure includes 211 residues so therefore only includes one domain, the receptor binding domain (RBD)
- The classification of domains was also verified using the scissors method which showed there were no points of flexible rotation and that secondary structures in both the ACE-2 domain and the Spike RBD held a globular structure.
- The ACE-2 protein has both alpha-helices and beta-sheets. The spike protein, however, only has beta-sheet secondary structures. This was determined by selecting the Ribbon view and surveying the two proteins for secondary structure.
Styles
- Below, see different styles of protein structure:
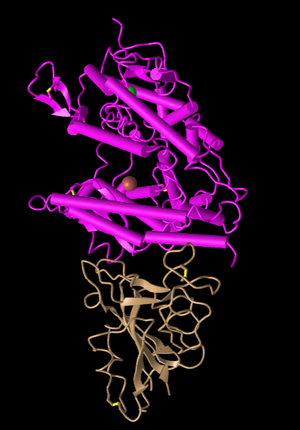
Figure 2: Plate and Cylinder Style. This style emphasizes the secondary structures with very unique plates and cylinder bars.
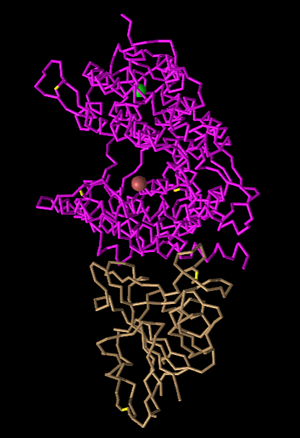
Figure 3: C-Alpha-Trace Style. This style is very similar to lines, however each residue is only one line.
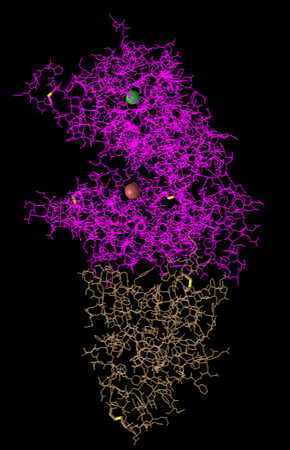
Figure 4: Lines Style. Side chain structures are shown in line formation.

Figure 5: Ball and Stick Style. Side chain structures are shown with each atom as a ball and their bonds as a line.
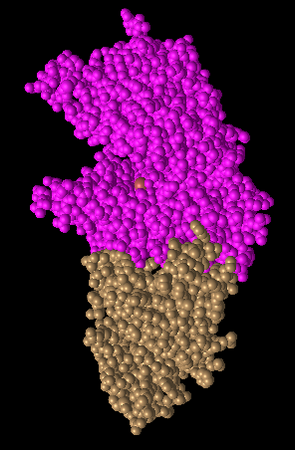
Figure 6: Spheres Style. Each amino acid is a sphere. This shows very globular forms.
Colors
- Below, see the different protein coloring options.
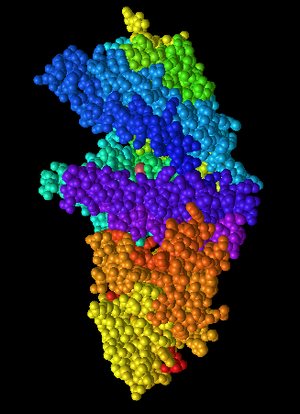
Figure 7: Spectrum coloring. Multiple colors are used. All residues are colored. It could be an attempt at coloring domains or groups of secondary structures.
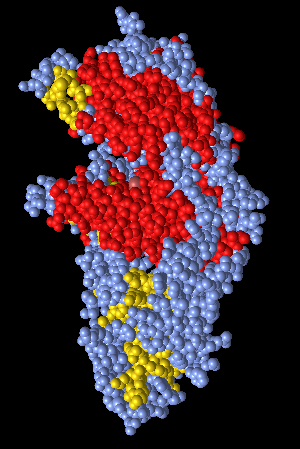
Figure 8: Secondary coloring. Secondary structures are colored either red (alpha-helices) or yellow (beta-sheets).
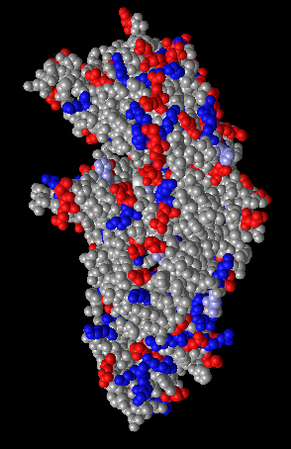
Figure 9: Charge coloring. Charged side chains are colored red or blue.
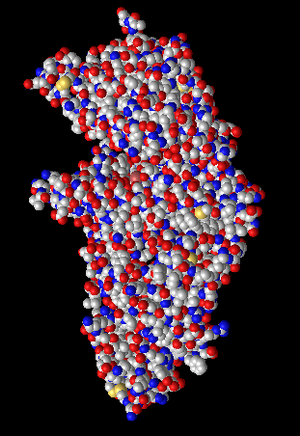
Figure 10: Atom coloring. Atoms of focus are colored. Red atoms are oxygen and blue are nitrogen.
Terminals
- N-terminus and C-terminus were located for both ACE-2 and Spike protein. This was done by viewing the proteins in ribbon style and following the direction of the arrows in the beta-sheets to find the C-terminus. This direction was reversed to locate the N-terminus.
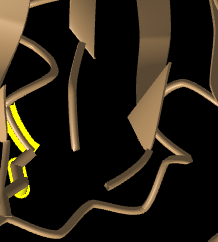
Figure 11: N-terminus (left) and C-terminus (right) of spike protein. Beta sheet can be seen pointing toward C-terminus.
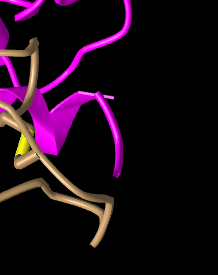
Figure 12: N terminus of ACE-2 protein (pink).
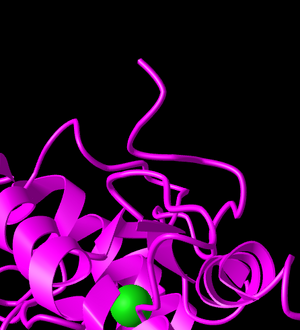
Figure 13: C terminus of ACE-2 protein.
Civet ACE2-Spike Protein: Figure 4B of Wan et al 2020
- Then, side chain interactions in the civet ACE2-spike protein structure were analyzed to recreate Figure 4B from Wan et al 2020.
- In the Analysis drop-down tab, "View Sequences and Annotations" was clicked to reveal the window. The details tab was clicked to show the sequence
- I then focused on the region of focus in Figure 4B of Wan et. al 2020. The pink and tan proteins were used for this focus. This region of focus is the receptor binding motif.
- The following residues were selected in the detail sequence window to be highlighted in yellow
- T31
- E35
- E38
- T82
- K353
- These residues were switched to the ball and stick style through the style drop-down tab. Because these residues were selected, they were the only ones to change style. They then were switched to the atom color.
- This process was repeated with the following residues of the spike protein sequence (3SCK_E)
- T487
- R479
- G480
- Y442
- P472
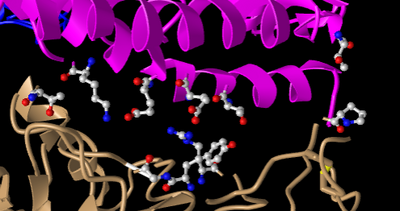
Figure 14: Replicate of Figure 4B from Wan et al 2020 without interactions. Focused on spike protein RBM area of interaction.
- E35 on ACE2 and R479 on the spike protein make an ionic bond with each other. Judging by the N groups on R479, it can be seen that R497 is the basic residue in this interaction. E35 on ACE2 has red atoms exposed, showing the O groups that mark it as the acidic residue in this interaction.
- T31 on ACE 2 and Y442 on the spike protein form a hydrogen bond. Both of these side chains belong to the uncharged polar group. This can be seen in Figure through the oxygen and nitrogen group located on T31 and the sole oxygen group located off the aromatic ring on Y442.
- Interactions were highlighted using the following methdods
- With the residues of focus still highlighted, I selected H-Bonds and Interactions in the Analysis drop-down menu.
- A new window appeared and in part 1 of this window, I unchecked the "Contacts and Interactions" box
- In part 2, I selected 3SCK_A
- In part 3, selected 3SCK_E
- In part 4, clicked 3D Display Interactions
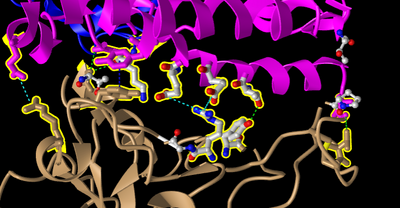
Figure 15: Replicate of Figure 4B from Wan et al 2020 with interactions. Focused on spike protein RBM area of interaction.
Beginning the Research Project
- What question will you answer about sequence-->structure-->function relationships in the spike and/or ACE2 protein?
- To what extent are mutations in the human spike protein related to geography and time?
- What sequences will you use?
- China: NC_045512 (collected December 2019)
- Spain: MT956913 (collected April 15th)
- Italy: MT_483879 (collected March 18th)
- Washington, USA: MT598633 (collected February 20th)
- India: MT940464 (collected July 27th)
- New Zealand: MT706050 (collected March 21st)
- China: MT911467 (collected August 14th)
- Peru: MW030279 (collected May 6th)
Scientific Conclusion
The NCBI 3D protein structure web application was successfully explored and used to achieve the goals of this lab. Different styles and color schemes revealed different aspects of protein structure. Finally, this knowledge was used to recreate Figure 4B from Wan et al, successfully mapping the interactions between the critical residues in the receptor binding motif of the civet ACE-2 binding spike protein.
Acknowledgements
- Consultation with Owen Dailey on amount of domains present in the NCBI database structure for Spike protein optimized for human ACE-2.
- Collaboration with Owen Dailey on the formation of research question.
- Zoom meeting with Owen Dailey to choose sequences to use in research project.
- Nida Patel helped me comment out text.
- Syntax copied from BIOL368/F20:Week_5 for civet spike protein 3d structure link
- Syntax for research project portion copied from Owen_R._Dailey_Week_5
- Background information used to understand protein secondary and tertiary structure as well as side chain properties found at Molecular Biology of the Cell. 4th edition.
References
- BIOL368/F20:Week_5
- iCn3D: Web-based 3D Structure Viewer 3SCI. (2020). Accessed Oct 1 and 7 2020. https://www.ncbi.nlm.nih.gov/Structure/icn3d/full.html?&mmdbid=97063&bu=1&showanno=1&source=full-feature
- iCn3D: Web-based 3D Structure Viewer 3SCK. Accessed Oct 7 2020. https://www.ncbi.nlm.nih.gov/Structure/icn3d/full.html?pdbid=%203SCK
- Wan, Y., Shang, J., Graham, R., Baric, R. S., & Li, F. (2020). Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. Journal of virology, 94(7).