Ian R. Wright Week 4
Ian Wright's Bioinformatics Portfolio
Assignment Pages
Individual Journal Entries
- Ian R. Wright Week 1
- Ian R. Wright Week 2
- Ian R. Wright Week 3
- Ian R. Wright Week 4
- Ian R. Wright Week 5
- Ian R. Wright Week 6
- Ian R. Wright Week 7
- Therapeutic Target Database (TTD) Review
- Ian R. Wright Week 9
- Ian R. Wright Week 10
- Ian R. Wright Week 11
- The D614G Research Group Week 12
- Ian R. Wright Week 14
- The D614G Research Group Week 14
Class Journals
BIOL368/F20:Class_Journal_Week_1
BIOL368/F20:Class_Journal_Week_2
BIOL368/F20:Class_Journal_Week_3
BIOL368/F20:Class_Journal_Week_4
BIOL368/F20:Class_Journal_Week_5
BIOL368/F20:Class_Journal_Week_6
BIOL368/F20:Class_Journal_Week_7
BIOL368/F20:Class_Journal_Week_8
BIOL368/F20:Class_Journal_Week_9
BIOL368/F20:Class_Journal_Week_10
BIOL368/F20:Class_Journal_Week_11
BIOL368/F20:Class_Journal_Week_12
BIOL368/F20:Class_Journal_Week_14
Purpose
The purpose of this exercise is to create a phylogenetic tree of 15 spike proteins from beta-coronaviruses. The spike protein sequences will then be aligned and assessed for divergence including a comparison of alignment with the phylogram. Both phylogram and sequence alignment will be compared with those provided in Wan et al 2020. Ultimately, this study is aimed at assessing the relatedness of SARS-CoV-2 spike proteins to other beta-coronavirus spike proteins.
Combined Methods and Results
Part 1: GenBank
- In the "Data and Tools" section of BIOL368/F20:Week_4, I clicked the sequence link for GenBank accession number MN908947.
- This GenBank record provides the name of the source organism (SARS-CoV-2), information regarding the source publication, and sequence data for the organism:
- Amino Acid sequence for each known locus
- Full nucleotide sequence of organism genome
- This GenBank record provides the name of the source organism (SARS-CoV-2), information regarding the source publication, and sequence data for the organism:
- The full nucleotide sequence was downloaded
- Clicked the Send to link in the upper right of the page.
- Selected Complete Record, File as the Destination, and FASTA as the format.
- Clicked the Create File button.
- Before opening the file, I right clicked the file and selected Open With... then Other and finally selected Microsoft Word
- It is also helpful to check the Always Open With box
- For verification purposes, I opened the file to make sure it was in FASTA format
- This was verified by the greater than symbol (>) followed by the accession number and organism information which then was followed by the genome sequence
- I searched GenBank for the Spike Protein sequence of my assigned accession number
- MK211378 - Coronavirus BtRs-BetaCoV/YN2018D
- This accession number brought me to the complete genome of BtRs-BetaCoV/YN2018D. To find the Spike Protein sequence, I scrolled down to "/product="spike glycoprotein" /protein_id="QDF43835.1"" and clicked the hyperlink attached to the protein accession number
- Hyperlinks for full sequence and spike protein sequence were added to the "Data and Tools" section of BIOL368/F20:Week_4 among sequences from related coronaviruses collected by other BioInformatics students for crowdsourcing purposes
- Spike Protein amino acid sequence was downloaded to hard drive in FASTA format
- Sequence provided below
- Sequence was added to the talk page for BIOL368/F20:Week_4
>QDF43835.1 spike glycoprotein [Coronavirus BtRs-BetaCoV/YN2018D] MKVLIVLLCLGLVTAQDGCGHISTKPQPLLDKFSSSRRGVYYNDDIFRSDVLHLTQDYFLPFDTNLTRYL SFNMDSATKVYFDNPTLPFGDGIYFAATEKSNVVRGWIFGSTMDNTTQSAIIVNNSTHIIIRVCYFNLCK EPMYAISNEQHYKSWVYQNAYNCTYDRVEQSFQLDTAPQTGNFKDLREYVFKNKDGFLSVYNAYSPIDIP RGLPVGFSVLKPILKLPIGINITSFKVVMSMFSRTTSNFLPEVAAYFVGNLKYSTFMLNFNENGTITDAI DCAQNPLSELKCTIKNFNVSKGIYQTSNFRVSPTHEVIRFPNITNRCPFDKVFNASRFPNVYAWERTKIS DCVADYTVLYNSTSFSTFKCYGVSPSKLIDLCFTSVYADTFLIRSSEVRQVAPGETGVIADYNYKLPDDF TGCVIAWNTAKQDQGQYYYRSSRKTKLKPFERDLTSDENGVRTLSTYDFYPNVPIEYQATRVVVLSFELL NAPATVCGPKLSTGLVKNQCVNFNFNGLRGTGVLTDSSKRFQSFQQFGRDTSDFTDSVRDPQTLEILDIT PCSFGGVSVITPGTNASSEVAVLYQDVNCTDVPTAIRADQLTPAWRVYSTGINVFQTQAGCLIGAEHVNA SYECDIPIGAGICASYHTASTLRSVGQKSIVAYTMSLGAENSIAYANNSIAIPTNFSISVTTEVMPVSMS KTSVDCTMYICGDSQECSNLLLQYGSFCTQLNRALTGIAIEQDKNTQEVFAQVKQMYKTPAIKDFGGFNF SQILPDPSKPTKRSFIEDLLFNKVTLADAGFMKQYGECLGDINARDLICAQKFNGLTVLPPLLTDDMIAA YTAALVSGTATAGWTFGAGAALQIPFAMQMAYRFNGIGVTQNVLYENQKQIANQFNKAISQIQESLTTTS TALGKLQDVVNQNAQALNTLVKQLSSNFGAISSVLNDILSRLDKVEAEVQIDRLITGRLQSLQTYVTQQL IRAAEIRASANLAATKMSECVLGQSKRVDFCGRGYHLMSFPQAAPHGVVFLHVTYVPSQEKNFTTAPAIC HEGKAYFPREGVFVSNGTSWFITQRNFYSPQIITTDNTFVAGSCDVVIGIINNTVYDPLQPELDSFKEEL DKYFKNHTSPDVDLGDISGINASVVNIQKEIDRLNEVAKNLNESLIDLQELGKYEQYIKWPWYVWLGFIA GLIAIVMATILLCCMTSCCSCLKGACSCGSCCKFDEDDSEPVLKGVKLHYT
Part 2: Phylogenetic Tree with Phylogeny.fr
- I went to the website www.phylogeny.fr and scrolled down to the section labeled ‘Phylogeny analysis’, and clicked on the text ‘One Click’
- I copied/pasted the list of sequences from the talk page to the 'or paste it here' section on the page and then clicked 'Submit'
- After rendering was completed, I clicked on the tab for '3. Alignment' and at the bottom of this page, clicked on 'Alignment in Clustal Format' under 'Outputs:'
- Sequence Alignment copied into 'CLUSTAL FORMAT: MUSCLE (3.8) multiple sequence alignment' section below
- Then I clicked on the Phylogenetic Tree tab
- I edited the Phylogenetic Tree using the 'Flip' function to place 'SARS Coronavirus Urbani' (Human infecting SARS-CoV-1 from 2002) directly next to 'Severe Acute Respiratory Syndrome' (SARS-CoV-2) for ease of comparison (Figure 1).
- saved the image to upload below sequence
- Sequence alignment was then compared to phylogenetic tree:
- Measurement via ruler and scale provided on tree allowed for the calculation of divergence percentage.
- Upon surface level visual inspection of alignment, it can be seen that in multiple instances, accession number QHD43416.1 (the spike protein for SARS-CoV-2) has drastic differences from other sequences. SARS-CoV-2 is located in a distant clade within the phylogenetic tree (Figure 1). AVP78031.1 (Bat-SARS-like coronavirus) is the other protein within this clade and even then, they are distantly related. Just between these two species of coronavirus, there is a ~17.5% divergence.
- Regarding the outgroups (AFS88936.1 and YP_0010399) (Figure 1), they seem well chosen due to drastic differences seen within the sequence alignment.
- When comparing SARS-CoV-1 to SARS-CoV-2 in the phylogenetic tree, it can be seen that there is a roughly 23.6% divergence in spike protein sequence (Figure 1). This is also reflected in the sequence alignment
- Sequence for Receptor Binding Domain was then isolated to be compared to Figure 3 of Wan et al 2020. RBD sequence can be seen in the 'Receptor Binding Domain Sequence' section
- RBD sequence alignment is very similar to that of Wan et al 2020, however, there are some differences:
- Not all residues are listed in the alignment from phylogeny.fr
- RBM is not highlighted in magenta in the Phy.fr alignment
- Critical residues are not highlighted in blue in Phy.fr alignment
- Sequences from a greater number of coronaviruses are included in the Phy.fr alignment
- There is a lesser amount of * fully conserved locations
- Most likely due to variety of species sequences included
- RBD sequence alignment is very similar to that of Wan et al 2020, however, there are some differences:
- Phylogenetic Tree was compared to the Tree (Figure 2) from Wan et al 2020
- The divergence scale in Wan et al 2020 is much smaller in magnitude but not size, allowing for more horizontal space to be used. This is helpful for visualization and divergence measurements.
- The Phylogenetic Tree from phylogeny.fr has a clade arrangement that allows for easy comparison between the viruses being studied in Wan et al 2020. In other words, SARS-CoV-1 and SARS-CoV-2 are closer
- SARS-CoV-2 (called 2019-nCoV in Wan et al 2020) in both phylogenetic trees shows high divergence from both SARS-CoV-1 and from the rest of the considered spike protein sequences
- SARS-CoV-1 shows high relatedness to sister groups in both trees
- In Wan et al 2020 Figure 2, there is another clade with two species in a clade with SARS-CoV-2 but not in my tree. This brought to attention that the accession numbers provided in the Wan et al 2020 tree are the accession numbers for the viruses themselves, not the spike protein numbers like in my phylogeny.
- In terms of reproducibility of Wan et al 2020, it is possible to reproduce the phylogeny and sequence alignment, however not possible to reproduce the atomic analysis of mutation effects on ACE-2 binding nor is it possible to recreate the optimized-for-binding spike protein sequence.
- Radial Phylograms can be reproduced using Geneious Prime with sequences from GenBank as described in materials and methods.
- Sequence alignment can be reproduced using Clustal Omega as described in materials and methods.
- There is no methodology listed for how atomic analysis was conducted. There was also no methodology for the creation of the theoretical optimized spike protein.
CLUSTAL FORMAT: MUSCLE (3.8) multiple sequence alignment
Markings below each column represent a degree of conservation across sequences: '*' for invariant, ':'for highly conserved, '.' for weakly conserved, and a space for not conserved
QDF43825.1 ---------MKLLVLV-----FATLVSSYTIEKCTDFD------DRTPPSNTQFLSSHRG
AGZ48818.1 ---------MKLLVLV-----FATLVSSYTIEKCLDFD------DRTPPANTQFLSSHRG
ALK02457.1 ----------MFIFLF-----FLTLTSGSDLESCTTFD------DVQAPNYPQHSSSRRG
AAS10463.1 ----------MFIFLL-----FLTLTSGSDLDRCTTFD------DVQAPNYTQHTSSMRG
AAP13441.1 ----------MFIFLL-----FLTLTSGSDLDRCTTFD------DVQAPNYTQHTSSMRG
AAP13567.1 ----------MFIFLL-----FLTLTSGSDLDRCTTFD------DVQAPNYTQHTSSMRG
QHD43416.1 ----------MFVFLV-----LLPLVSSQ----CVNLT------TRTQLPPAYTNSFTRG
AVP78031.1 -----------MLFFL-----FLQFALVN--SQCVNLT------GRTPLNPNYTNSSQRG
ABD75323.1 --------MKILIFAF-----LVTLVKAQ--EGCGVIN------LRTQPKLTQVSSSRRG
QDF43835.1 --------MKVLIVLL-----CLGLVTAQ--DGCGHIS------TKPQPLLDKFSSSRRG
ABD75332.1 --------MKVLIFAL-----LFSLAKAQ--EGCGIIS------RKPQPKMEKVSSSRRG
QDF43820.1 --------MKILIFAF-----LVTLVEAQ--EGCGIIS------RKPQPKMAQVSSSRRG
AAZ67052.1 --------MKILILAF-----LASLAKAQ--EGCGIIS------RKPQPKMAQVSSSRRG
AFS88936.1 ----MIHSVFLLMFLLTPTESYVDVGPDSVKSACIEVDIQQTFFDKTWPRPIDVSKA-DG
YP_0010399 MTLLMCLLMSLLIFVRGCDSQFVDMSPASNTSECLESQVDAAAFSKLMWPYPIDPSKVDG
::. . * . *
QDF43825.1 VYYPDDIFRSNVLHLVQDHFLPFDSNVTRFITFGLN-------------FDN---PIIPF
AGZ48818.1 VYYPDDIFRSNVLHLVQDHFLPFDSNVTRFITFGLN-------------FDN---PIIPF
ALK02457.1 VYYPDEIFRSDTLYLTQDLFLPFYSNVTGFHTINHR-------------FDN---PVIPF
AAS10463.1 VYYPDEIFRSDTLYLTQDLFLPFYSNVTGFHTINHT-------------FDD---PVIPF
AAP13441.1 VYYPDEIFRSDTLYLTQDLFLPFYSNVTGFHTINHT-------------FGN---PVIPF
AAP13567.1 VYYPDEIFRSDTLYLTQDLFLPFYSNVTGFHTINHT-------------FDN---PVIPF
QHD43416.1 VYYPDKVFRSSVLHSTQDLFLPFFSNVTWFHAIHVS------GTNGTKRFDN---PVLPF
AVP78031.1 VYYPDTIYRSDTLVLSQGYFLPFYSNVSWYYSLTTN-------NAATKRTDN---PILDF
ABD75323.1 VYYNDDIFRSDVLHLTQDYFLPFHSNLTQYFSLNIE-------SDKIVYFDN---PILKF
QDF43835.1 VYYNDDIFRSDVLHLTQDYFLPFDTNLTRYLSFNMD-------SATKVYFDN---PTLPF
ABD75332.1 VYYNDDIFRSDVLHLTQDYFLPFDSNLTQYFSLNID-------SNKYTYFDN---PILDF
QDF43820.1 VYYNDDIFRSDVLHLTQDYFLPFDSNLTQYFSLNVD-------SDRYTYFDN---PILDF
AAZ67052.1 VYYNDDIFRSNVLHLTQDYFLPFDSNLTQYFSLNVD-------SDRFTYFDN---PILDF
AFS88936.1 IIYPQGRTYSNITITYQGLF-PYQGDHGDMYVYSAG--HATGTTPQKLFVANYSQDVKQF
YP_0010399 IIYPLGRTYSNITLAYTGLF-PLQGDLGSQYLYSVSHAVGHDGDPTKAYISNYSLLVNDF
: * *. . * * : : *
QDF43825.1 RDGVYF----AATEKSNVIRG-------------WVFGSTMNNKSQ---------SVIIM
AGZ48818.1 KDGIYF----AATEKSNVIRG-------------WVFGSTMNNKSQ---------SVIIM
ALK02457.1 KDGVYF----AATEKSNVVRG-------------WVFGSTMNNKSQ---------SVIII
AAS10463.1 KDGIYF----AATEKSNVVRG-------------WVFGSTMNNKSQ---------SVIII
AAP13441.1 KDGIYF----AATEKSNVVRG-------------WVFGSTMNNKSQ---------SVIII
AAP13567.1 KDGIYF----AATEKSNVVRG-------------WVFGSTMNNKSQ---------SVIII
QHD43416.1 NDGVYF----ASTEKSNIIRG-------------WIFGTTLDSKTQ---------SLLIV
AVP78031.1 KDGIYF----AATEHSNIIRG-------------WIFGTTLDNTSQ---------SLLIV
ABD75323.1 GDGVYF----AATEKSNVIRG-------------WVFGSTFDNTTQ---------SAIIV
QDF43835.1 GDGIYF----AATEKSNVVRG-------------WIFGSTMDNTTQ---------SAIIV
ABD75332.1 GDGVYF----AATEKSNVIRG-------------WIFGSSFDNTTQ---------SAIIV
QDF43820.1 GDGVYF----AATEKSNVIRG-------------WIFGSTFDNTTQ---------SAVIV
AAZ67052.1 GDGVYF----AATEKSNVIRG-------------WIFGSTFDNTTQ---------SAVIV
AFS88936.1 ANGFVVRIGAAANSTGTVIISPSTSATIRKIYPAFMLGSSVGNFSDGKMGRFFNHTLVLL
YP_0010399 DNGFVVRIGAAANSTGTIVISPSVNTKIKKAYPAFILGSSLTNTSAGQ-PLYANYSLTII
:*. . *:.. ..:: . :::*::. . : : ::
QDF43825.1 NNSTNLVIRACNFELCDNPFFVVLRSNNTQIPSY------IFNNAFN-CTFEYVSKDFNL
AGZ48818.1 NNSTNLVIRACNFELCDNPFFVVLKSNNTQIPSY------IFNNAFN-CTFEYVSKDFNL
ALK02457.1 NNSTNVVIRACNFELCDNPFFAVSKPTGTQTHTM------IFDNAFN-CTFEYISDSFSL
AAS10463.1 NNSTNVVIRACNFELCDNPFFVVSKPMGTRTHTM------IFDNAFN-CTFEYISDAFSL
AAP13441.1 NNSTNVVIRACNFELCDNPFFAVSKPMGTQTHTM------IFDNAFN-CTFEYISDAFSL
AAP13567.1 NNSTNVVIRACNFELCDNPFFAVSKPMGTQTHTM------IFDNAFN-CTFEYISDAFSL
QHD43416.1 NNATNVVIKVCEFQFCNDPFLGVYY--HKNNKSWMESEFRVYSSANN-CTFEYVSQPFLM
AVP78031.1 NNATNVIIKVCNFDFCYDP-YLSGY--YHNNKTWSIREFAVYSSYAN-CTFEYVSKSFML
ABD75323.1 NNSTHIIIRVCYFNLCKDPMYTVSA--GTQKSSW------VYQSAFN-CTYDRVEKSFQL
QDF43835.1 NNSTHIIIRVCYFNLCKEPMYAISN--EQHYKSW------VYQNAYN-CTYDRVEQSFQL
ABD75332.1 NNSTHIIIRVCNFNLCKEPMYTVSK--GTQQSSW------VYQSAFN-CTYDRVEKSFQL
QDF43820.1 NNSTHIIIRVCNFNLCKEPMYTVSR--GTQQSSW------VYQSAFN-CTYDRVERSFQL
AAZ67052.1 NNSTHIIIRVCNFNLCKEPMYTVSR--GAQQSSW------VYQSAFN-CTYDRVEKSFQL
AFS88936.1 PDGCGTLLRAFYCIL--EPRSGNHCPAGNSYTSF-----ATYHTPATDCSDGNYNRNASL
YP_0010399 PDGCGTVLHAFYCIL--KPRTVNRCPSGTGYVSY-----FIYETVHNDCQ-STINRNASL
:. ::.. : .* : : . . * . :
QDF43825.1 DIGEKPGNFKDLREFVFRNKDG--------FLHVYSGYQPISAASGLPTGF--NALKPIF
AGZ48818.1 DLGEKPGNFKDLREFVFRNKDG--------FLHVYSGYQPISAASGLPTGF--NALKPIF
ALK02457.1 DVAEKSGNFKHLREFVFKNKDG--------FLYVYKGYQPIDVVRDLPSGF--NILKPIF
AAS10463.1 DVSEKSGNFKHLREFVFKNKDG--------FLYVYKGYQPIDVVRDLPSGF--NTLKPIF
AAP13441.1 DVSEKSGNFKHLREFVFKNKDG--------FLYVYKGYQPIDVVRDLPSGF--NTLKPIF
AAP13567.1 DVSEKSGNFKHLREFVFKNKDG--------FLYVYKGYQPIDVVRDLPSGF--NTLKPIF
QHD43416.1 DLEGKQGNFKNLREFVFKNIDG--------YFKIYSKHTPINLVRDLPQGF--SALEPLV
AVP78031.1 NISGNGGLFNTLREFVFRNVDG--------HFKIYSKFTPVNLNRGLPTGL--SVLQPLV
ABD75323.1 DTSPKTGNFTDLREFVFKNRDG--------FFTAYQTYTPVNLLRGLPSGL--SVLKPIL
QDF43835.1 DTAPQTGNFKDLREYVFKNKDG--------FLSVYNAYSPIDIPRGLPVGF--SVLKPIL
ABD75332.1 DTAPKTGNFKDLREYVFKNKGG--------FLRVYQTYTAVNLPRGFPAGF--SVLRPIL
QDF43820.1 DTAPKTGNFKDLREYVFKNRDG--------FLSVYQTYTAVNLPRGLPIGF--SVLRPIL
AAZ67052.1 DTAPKTGNFKDLREYVFKNRDG--------FLSVYQTYTAVNLPRGLPIGF--SVLRPIL
AFS88936.1 NSFKE---YFNLRNCTFMYTYNITEDEILEWFGITQTAQGVHLFSSRYVDLYGGNMFQFA
YP_0010399 NSFK---SFFDLVNCTFFNSWDITADETKEWFGITQDTQGVHLYSSRKGDLYGGNMFRFA
: : * : .* . : . : . .: . : :
QDF43825.1 KLPLGINITNFRTLLTAF------PPNPGYWGTSAAAYFVGYLKPTTFMLKYDENGTITD
AGZ48818.1 KLPLGINITNFRTLLTAF------PPRPDYWGTSAAAYFVGYLKPTTFMLKYDENGTITD
ALK02457.1 KLPLGINITNFRAILTAF------LPAQDTWGTSAAAYFVGYLKPATFMLKYDENGTITD
AAS10463.1 KLPLGINITNFRAILTAF------SPAQDTWGTSAAAYFVGYLKPTTFMLKYDENGTITD
AAP13441.1 KLPLGINITNFRAILTAF------SPAQDIWGTSAAAYFVGYLKPTTFMLKYDENGTITD
AAP13567.1 KLPLGINITNFRAILTAF------SPAQDTWGTSAAAYFVGYLKPTTFMLKYDENGTITD
QHD43416.1 DLPIGINITRFQTLLALHRSYLTPGDSSSGWTAGAAAYYVGYLQPRTFLLKYNENGTITD
AVP78031.1 ELPVSINITKFRTLLTIHRGD---PMPNNGWTAFSAAYFVGYLKPRTFMLKYNENGTITD
ABD75323.1 KLPFGINITSFRVVMAMF------SKTTSNYVPESAAYYVGNLKQSTFMLSFNQNGTIVD
QDF43835.1 KLPIGINITSFKVVMSMF------SRTTSNFLPEVAAYFVGNLKYSTFMLNFNENGTITD
ABD75332.1 KLPFGINITSYRVVMTMF------SQFNSNFLPESAAYYVGNLKYTTFMLSFNENGTITD
QDF43820.1 KLPFGINITSYRVVMAMF------SQTTSNFLPESAAYYVGNLKYTTFMLRFNENGTITD
AAZ67052.1 KLPFGINITSYRVVMAMF------SQTTSNFLPESAAYYVGNLKYTTFMLSFNENGTITN
AFS88936.1 TLPVYDTIKYYSIIPHSIRSI---QSDRKAW----AAFYVYKLQPLTFLLDFSVDGYIRR
YP_0010399 TLPVYEGIKYYTVIPRSFRSK---ANKREAW----AAFYVYKLHQLTYLLDFSVDGYIRR
**. *. : : : **::* *: *::* :. :* *
QDF43825.1 AVDCSQNPLAELKCSVKSFEIDKGIYQTSNFRVAPSKEVVRFPNITNLCPFGEVFNATTF
AGZ48818.1 AVDCSQNPLAELKCSVKSFEIDKGIYQTSNFRVAPSKEVVRFPNITNLCPFGEVFNATTF
ALK02457.1 AVDCSQNPLAELKCSVKSFEIDKGIYQTSNFRVAPSKEVVRFPNITNLCPFGEVFNATTF
AAS10463.1 AVDCSQNPLAELKCSVKSFEIDKGIYQTSNFRVVPSGDVVRFPNITNLCPFGEVFNATKF
AAP13441.1 AVDCSQNPLAELKCSVKSFEIDKGIYQTSNFRVVPSGDVVRFPNITNLCPFGEVFNATKF
AAP13567.1 AVDCSQNPLAELKCSVKSFEIDKGIYQTSNFRVVPSGDVVRFPNITNLCPFGEVFNATKF
QHD43416.1 AVDCALDPLSETKCTLKSFTVEKGIYQTSNFRVQPTESIVRFPNITNLCPFGEVFNATRF
AVP78031.1 AVDCALDPLSETKCTLKSLTVQKGIYQTSNFRVQPTQSVVRFPNITNVCPFHKVFNATRF
ABD75323.1 AVDCSQDPLAELKCTTKSFNVSKGIYQTSNFRVSPVTEVVRFPNITNLCPFDKVFNATRF
QDF43835.1 AIDCAQNPLSELKCTIKNFNVSKGIYQTSNFRVSPTHEVIRFPNITNRCPFDKVFNASRF
ABD75332.1 AVDCSQNPLAELKCTIKNFNVSKGIYQTSNFRVTPTQEVVRFPNITNRCPFDKVFNASRF
QDF43820.1 AIDCAQNPLAELKCTIKNFNVSKGIYQTSNFRVSPTQEVVRFPNITNRCPFDKVFNASRF
AAZ67052.1 AIDCAQNPLAELKCTIKNFNVSKGIYQTSNFRVSPTQEVIRFPNITNRCPFDKVFNATRF
AFS88936.1 AIDCGFNDLSQLHCSYESFDVESGVYSVSSFEAKPSGSVVEQAEGVE-CDFSPLLSGTP-
YP_0010399 AIDCGHDDLSQLHCSYTSFEVDTGVYSVSSYEASATGTFIEQPNATE-CDFSPMLTGVA-
*:**. : *:: :*: .: :..*:*..*.: . . .: .: .: * * ::..
QDF43825.1 PSVYAWERKRISNCVADYSVLYNSTSFSTFKCYGVSATKLNDLCFSNVYADSFVVKGDDV
AGZ48818.1 PSVYAWERKRISNCVADYSVLYNSTSFSTFKCYGVSATKLNDLCFSNVYADSFVVKGDDV
ALK02457.1 PSVYAWERKRISNCVADYSVLYNSTSFSTFKCYGVSATKLNDLCFSNVYADSFVVKGDDV
AAS10463.1 PSVYAWERKRISNCVADYSVLYNSTSFSTFKCYGVSATKLNDLCFSNVYADSFVVKGDDV
AAP13441.1 PSVYAWERKKISNCVADYSVLYNSTFFSTFKCYGVSATKLNDLCFSNVYADSFVVKGDDV
AAP13567.1 PSVYAWERKKISNCVADYSVLYNSTFFSTFKCYGVSATKLNDLCFSNVYADSFVVKGDDV
QHD43416.1 ASVYAWNRKRISNCVADYSVLYNSASFSTFKCYGVSPTKLNDLCFTNVYADSFVIRGDEV
AVP78031.1 PSVYAWERTKISDCIADYTVFYNSTSFSTFKCYGVSPSKLIDLCFTSVYADTFLIRFSEV
ABD75323.1 PSVYAWERTKISDCVADYTVFYNSTSFSTFNCYGVSPSKLIDLCFTSVYADTFLIRFSEV
QDF43835.1 PNVYAWERTKISDCVADYTVLYNSTSFSTFKCYGVSPSKLIDLCFTSVYADTFLIRSSEV
ABD75332.1 PNVYAWERTKISDCVADYTVLYNSTSFSTFKCYGVSPSKLIDLCFTSVYADTFLIRSSEV
QDF43820.1 PNVYAWERTKISDCVADYTVLYNSTSFSTFKCYGVSPSKLIDLCFTSVYADTFLIRSSEV
AAZ67052.1 PNVYAWERTKISDCVADYTVLYNSTSFSTFKCYGVSPSKLIDLCFTSVYADTFLIRSSEV
AFS88936.1 PQVYNFKRLVFTNCNYNLTKLLSLFSVNDFTCSQISPAAIASNCYSSLILDYFSYPLSMK
YP_0010399 PQVYNFKRLVFSNCNYNLTKLLSLFAVDEFSCNGISPDSIARGCYSTLTVDYFAYPLSMK
..** ::* :::* : : : . .. *.* :*. : *::.: * * .
QDF43825.1 RQIAPGQTGVIADYNYKLPDDFMGC-VLAWNTRNIDATSTGNYNYKYRSLRHGKLRPFER
AGZ48818.1 RQIAPGQTGVIADYNYKLPDDFTGC-VLAWNTRNIDATQTGNYNYKYRSLRHGKLRPFER
ALK02457.1 RQIAPGQTGVIADYNYKLPDDFTGC-VLAWNTRNIDATQTGNYNYKYRSLRHGKLRPFER
AAS10463.1 RQIAPGQTGVIADYNYKLPDDFMGC-VLAWNTRNIDATSTGNYNYKYRYLRHGKLRPFER
AAP13441.1 RQIAPGQTGVIADYNYKLPDDFMGC-VLAWNTRNIDATSTGNYNYKYRYLRHGKLRPFER
AAP13567.1 RQIAPGQTGVIADYNYKLPDDFMGC-VLAWNTRNIDATSTGNYNYKYRYLRHGKLRPFER
QHD43416.1 RQIAPGQTGKIADYNYKLPDDFTGC-VIAWNSNNLDSKVGGNYNYLYRLFRKSNLKPFER
AVP78031.1 RQVAPGQTGVIADYNYKLPDDFTGC-VIAWNTAKQD---VGNYF--YRSHRSTKLKPFER
ABD75323.1 RQVAPGQTGVIADYNYKLPDDFTGC-VIAWNTAKQD---VGSYF--YRSHRSSKLKPFER
QDF43835.1 RQVAPGETGVIADYNYKLPDDFTGC-VIAWNTAKQD---QGQYY--YRSSRKTKLKPFER
ABD75332.1 RQVAPGETGVIADYNYKLPDDFTGC-VIAWNTAQQD---QGQYY--YRSYRKEKLKPFER
QDF43820.1 RQVAPGETGVIADYNYKLPDDFTGC-VIAWNTAKQD---TGHYY--YRSHRKTKLKPFER
AAZ67052.1 RQVAPGETGVIADYNYKLPDDFTGC-VIAWNTAKQD---QGQYY--YRSHRKTKLKPFER
AFS88936.1 SDLSVSSAGPISQFNYKQSFSNPTC-LILATVPHNLTTITKPLKYSYINKCSRLLSDDRT
YP_0010399 SYIRPGSAGNIPLYNYKQSFANPTCRVMASVLANVTITKPHAYG--YIS-KCSRLTGANQ
: ..:* *. :*** . * :: : * *
QDF43825.1 DISNVPFSPDGKPCTPP-AF-NCYW-----------PLNDYGFFTTNGIGYQPYRVVVLS
AGZ48818.1 DISNVPFSPDGKPCTPP-AF-NCYW-----------PLNDYGFYITNGIGYQPYRVVVLS
ALK02457.1 DISNVPFSPDGKPCTPP-AF-NCYW-----------PLNDYGFYITNGIGYQPYRVVVLS
AAS10463.1 DISNVPFSPDGKPCTPP-AP-NCYW-----------PLNGYGFYTTSGIGYQPYRVVVLS
AAP13441.1 DISNVPFSPDGKPCTPP-AL-NCYW-----------PLNDYGFYTTTGIGYQPYRVVVLS
AAP13567.1 DISNVPFSPDGKPCTPP-AL-NCYW-----------PLNDYGFYTTTGIGYQPYRVVVLS
QHD43416.1 DISTEIYQAGSTPCNGVEGF-NCYF-----------PLQSYGFQPTNGVGYQPYRVVVLS
AVP78031.1 DLSSDE---------------NGVR-----------TLSTYDFNPNVPLEYQATRVVVLS
ABD75323.1 DLSSEE---------------NGVR-----------TLSTYDFNQNVPLEYQATRVVVLS
QDF43835.1 DLTSDE---------------NGVR-----------TLSTYDFYPNVPIEYQATRVVVLS
ABD75332.1 DLSSDE---------------NGVY-----------TLSTYDFYPSIPVEYQATRVVVLS
QDF43820.1 DLSSDDG--------------NGVY-----------TLSTYDFNPNVPVAYQATRVVVLS
AAZ67052.1 DLSSDE---------------NGVR-----------TLSTYDFYPSVPVAYQATRVVVLS
AFS88936.1 EVPQLVNANQYSPCVSI-VP-STVWEDGDYYRKQLSPLEGGGWLVASGSTVAMTEQLQMG
YP_0010399 DVETPLYINPGEYSICRDFSPGGFSEDGQVFKRTLTQFEGGGLLIGVGTRVPMTDNLQMS
:: . :. . : :.
QDF43825.1 FELL----NAPATVC-----GPKLSTDLIKNQCVNFNFNGLTGTGVLTPSSKRFQPFQQF
AGZ48818.1 FELL----NAPATVC-----GPKLSTDLIKNQCVNFNFNGLTGTGVLTPSSKRFQPFQQF
ALK02457.1 FELL----NAPATVC-----GPKLSTDLIKNQCVNFNFNGLTGTGVLTPSSKRFQPFQQF
AAS10463.1 FELL----NAPATVC-----GPKLSTDLIKNQCVNFNFNGLTGTGVLTPSSKRFQPFQQF
AAP13441.1 FELL----NAPATVC-----GPKLSTDLIKNQCVNFNFNGLTGTGVLTPSSKRFQPFQQF
AAP13567.1 FELL----NAPATVC-----GPKLSTDLIKNQCVNFNFNGLTGTGVLTPSSKRFQPFQQF
QHD43416.1 FELL----HAPATVC-----GPKKSTNLVKNKCVNFNFNGLTGTGVLTESNKKFLPFQQF
AVP78031.1 FELL----NAPATVC-----GPKLSTQLVKNQCVNFNFNGLKGTGVLTDSSKRFQSFQQF
ABD75323.1 FELL----NAPATVC-----GPKLSTSLVKNQCVNFNFNGFKGTGVLTDSSKTFQSFQQF
QDF43835.1 FELL----NAPATVC-----GPKLSTGLVKNQCVNFNFNGLRGTGVLTDSSKRFQSFQQF
ABD75332.1 FELL----NAPATVC-----GPKLSTQLVKNQCVNFNFNGLRGTGVLTTSSKRFQSFQQF
QDF43820.1 FELL----NAPATVC-----GPKLSTQLVKNQCVNFNFNGLKGTGVLTDSSKRFQSFQQF
AAZ67052.1 FELL----NAPATVC-----GPKLSTQLVKNQCVNFNFNGLKGTGVLTESSKRFQSFQQF
AFS88936.1 FGITVQYGTDTNSVCPKLEFANDTKIASQLGNCVEYSLYGVSGRGVFQNCTAVGVRQQRF
YP_0010399 FIISVQYGTGTDSVCPMLDLGDSLTITNRLGKCVDYSLYGVTGRGVFQNCTAVGVKQQRF
* : . :** . . . .:**::.: *. * **: .. *.*
QDF43825.1 GRDVSD-FTDSVRDPKTSEILDISPCSFGGVSVITPGTNTSSEVAVLYQDVNCTDVPVAI
AGZ48818.1 GRDVSD-FTDSVRDPKTSEILDISPCSFGGVSVITPGTNTSSEVAVLYQDVNCTDVPVAI
ALK02457.1 GRDVLD-FTDSVRDPKTSEILDISPCSFGGVSVITPGTNTSSEVAVLYQDVNCTDVPVAI
AAS10463.1 GRDVSD-FTDSVRDPKTSEILDISPCSFGGVSVITPGTNASSEVAVLYQDVNCTDVSTLI
AAP13441.1 GRDVSD-FTDSVRDPKTSEILDISPCSFGGVSVITPGTNASSEVAVLYQDVNCTDVSTAI
AAP13567.1 GRDVSD-FTDSVRDPKTSEILDISPCSFGGVSVITPGTNASSEVAVLYQDVNCTDVSTAI
QHD43416.1 GRDIAD-TTDAVRDPQTLEILDITPCSFGGVSVITPGTNTSNQVAVLYQDVNCTEVPVAI
AVP78031.1 GKDASD-FIDSVRDPQTLEILDITPCSFGGVSVITPGTNTSLEVAVLYQDVNCTDVPTTI
ABD75323.1 GRDASD-FTDSVRDPQTLRILDISPCSFGGVSVITPGTNTSSAVAVLYQDVNCTDVPRTI
QDF43835.1 GRDTSD-FTDSVRDPQTLEILDITPCSFGGVSVITPGTNASSEVAVLYQDVNCTDVPTAI
ABD75332.1 GRDTSD-FTDSVRDPQTLEILDISPCSFGGVSVITPGTNASSEVAVLYQDVNCTDVPTSI
QDF43820.1 GRDTSD-FTDSVRDPQTLEILDITPCSFGGVSVITPGTNASSEVAVLYQDVNCTDVPTAI
AAZ67052.1 GRDTSD-FTDSVRDPQTLEILDISPCSFGGVSVITPGTNASSEVAVLYQDVNCTDVPAAI
AFS88936.1 VYDAYQNLVGYYSDDGNYYCLR--ACVSVPVSVIY--DKETKTHATLFGSVACEHISSTM
YP_0010399 VYDSFDNLVGYYSDDGNYYCVR--PCVSVPVSVIY--DKSTNLHATLFGSVACEHVTTMM
* : . * . : .* **** : : *.*: .* * :. :
QDF43825.1 --HADQLTPAWRIYSTGNNVFQTQAGCLIGAEHVDTSY---ECDIPIGAGICASYHTVSS
AGZ48818.1 --HADQLTPSWRVYSTGNNVFQTQAGCLIGAEHVDTSY---ECDIPIGAGICASYHTVSS
ALK02457.1 --HADQLTPSWRVYSTGNNVFQTQAGCLIGAEHVDTSY---ECDIPIGAGICASYHTVSS
AAS10463.1 --HAEQLTPAWRIYSTGNNVFQTQAGCLIGAEHVDTSY---ECDIPIGAGICASYHTVSS
AAP13441.1 --HADQLTPAWRIYSTGNNVFQTQAGCLIGAEHVDTSY---ECDIPIGAGICASYHTVSL
AAP13567.1 --HADQLTPAWRIYSTGNNVFQTQAGCLIGAEHVDTSY---ECDIPIGAGICASYHTVSL
QHD43416.1 --HADQLTPTWRVYSTGSNVFQTRAGCLIGAEHVNNSY---ECDIPIGAGICASYQTQTN
AVP78031.1 --HADQLTPAWRIYATGTNVFQTQAGCLIGAEHVNASY---ECDIPIGAGICASYHTASI
ABD75323.1 --QADQLAPSWRVYTTGPYVFQTQAGCLIGAEHVNASY---QCDIPIGAGICASYHTASH
QDF43835.1 --RADQLTPAWRVYSTGINVFQTQAGCLIGAEHVNASY---ECDIPIGAGICASYHTAST
ABD75332.1 --HADQLTPAWRVYSTGVNVFQTQAGCLIGAEHVNASY---ECDIPIGAGICASYHTASV
QDF43820.1 --RADQLTPAWRVYSTGVNVFQTQAGCLIGAEHVNASY---ECDIPIGAGICASYHTAST
AAZ67052.1 --HADQLTPAWRVYSTGTNVFQTQAGCLIGAEHVNASY---ECDIPIGAGICASYHTAST
AFS88936.1 SQYSRSTRSMLKRRDSTYGPLQTPVGCVLGL--VNSSLFVEDCKLPLGQSLCALPDTPST
YP_0010399 S-QFSRLTQSNLRRRDSNIPLQTAVGCVIGLS--NNSLVVSDCKLPLGQSLCAV-PPVST
:** .**::* : * :*.:*:* .:** . :
QDF43825.1 ----LRSTS----QKSI--------VAYTMSLGADSSIAYSNNTIAIPTNFSISITTEVM
AGZ48818.1 ----LRSTS----QKSI--------VAYTMSLGADSSIAYSNNTIAIPTNFSISITTEVM
ALK02457.1 ----LRSTS----QKSI--------VAYTMSLGADSSIAYSNNTIAIPTNFSISITTEVM
AAS10463.1 ----LRSTS----QKSI--------VAYTMSLGADSSIAYSNNTIAIPTNFSISITTEVM
AAP13441.1 ----LRSTS----QKSI--------VAYTMSLGADSSIAYSNNTIAIPTNFSISITTEVM
AAP13567.1 ----LRSTS----QKSI--------VAYTMSLGADSSIAYSNNTIAIPTNFSISITTEVM
QHD43416.1 SPRRARSVA----SQSI--------IAYTMSLGAENSVAYSNNSIAIPTNFTISVTTEIL
AVP78031.1 ----LRSTS----QKAI--------VAYTMSLGAENSIAYANNSIAIPTNFSISVTTEVM
ABD75323.1 ----LRSTG----QKSI--------VAYTMSLGAENSVAYANNSIAIPTNFSISVTTEVM
QDF43835.1 ----LRSVG----QKSI--------VAYTMSLGAENSIAYANNSIAIPTNFSISVTTEVM
ABD75332.1 ----LRSTG----QKSI--------VAYTMSLGAENSIAYANNSIAIPTNFSISVTTEVM
QDF43820.1 ----LRSVG----QKSI--------VAYTMSLGAENSIAYANNSIAIPTNFSISVTTEVM
AAZ67052.1 ----LRSVG----QKSI--------VAYTMSLGAENSIAYANNSIAIPTNFSISVTTEVM
AFS88936.1 ----LTPRS----VRSVPGEMRLASIAFNHPIQVDQ-LNSSYFKLSIPTNFSFGVTQEYI
YP_0010399 ----FRSYSASQFQLAV--------LNYTSPIVV-TPINSSGFTAAIPTNFSFSVTQEYI
. . :: : :. .: . : : . :*****::.:* * :
QDF43825.1 PVSMAKTSVDCNMYICGDSTECANLLLQYGSFCTQLNRALSGIAVEQDRNTREVFAQVKQ
AGZ48818.1 PVSMAKTSVDCNMYICGDSTECANLLLQYGSFCTQLNRALSGIAVEQDRNTREVFAQVKQ
ALK02457.1 PVSMAKTSVDCNMYICGDSTECANLLLQYGSFCTQLNRALSGIAVEQDRNTREVFAQVKQ
AAS10463.1 PVSMAKTSVDCNMYICGDSTECANLLLQYGSFCRQLNRALSGIAAEQDRNTREVFVQVKQ
AAP13441.1 PVSMAKTSVDCNMYICGDSTECANLLLQYGSFCTQLNRALSGIAAEQDRNTREVFAQVKQ
AAP13567.1 PVSMAKTSVDCNMYICGDSTECANLLLQYGSFCTQLNRALSGIAAEQDRNTREVFAQVKQ
QHD43416.1 PVSMTKTSVDCTMYICGDSTECSNLLLQYGSFCTQLNRALTGIAVEQDKNTQEVFAQVKQ
AVP78031.1 PVSMAKTSVDCTMYICGDSIECSNLLLQYGSFCTQLNRALSGIAIEQDKNTQEVFAQVKQ
ABD75323.1 PVSMAKTSVDCTMYICGDSLECSNLLLQYGSFCTQLNRALSGIAVEQDKNTQEVFAQVKQ
QDF43835.1 PVSMSKTSVDCTMYICGDSQECSNLLLQYGSFCTQLNRALTGIAIEQDKNTQEVFAQVKQ
ABD75332.1 PVSIAKTSVDCTMYICGDSLECSNLLLQYGSFCTQLNRALTGIAIEQDKNTQEVFAQVKQ
QDF43820.1 PVSMAKTSVDCTMYICGDSQECSNLLLQYGSFCTQLNRALTGVALEQDKNTQEVFAQVKQ
AAZ67052.1 PVSMAKTSVDCTMYICGDSLECSNLLLQYGSFCTQLNRALSGIAIEQDKNTQEVFAQVKQ
AFS88936.1 QTTIQKVTVDCKQYVCNGFQKCEQLLREYGQFCSKINQALHGANLRQDDSVRNLFASVKS
YP_0010399 ETSIQKVTVDCKQYVCNGFTRCEKLLVEYGQFCSKINQALHGANLRQDESVYSLYSNIKT
.:: *.:***. *:*.. * :** :**.** ::*.** * ** .. .:: .:*
QDF43825.1 MYKTPTLKD-FGG-FNFSQILPDPLKPTKRSF---IEDLLFNKVTLADAGFMKQYGECL-
AGZ48818.1 MYKTPTLKD-FGG-FNFSQILPDPLKPTKRSF---IEDLLFNKVTLADAGFMKQYGECL-
ALK02457.1 MYKTPTLKD-FGG-FNFSQILPDPLKPTKRSF---IEDLLFNKVTLADAGFMKQYGECL-
AAS10463.1 MYKTPTLKD-FGG-FNFSQILPDPLKPTKRSF---IEDLLFNKVTLADAGFMKQYGECL-
AAP13441.1 MYKTPTLKY-FGG-FNFSQILPDPLKPTKRSF---IEDLLFNKVTLADAGFMKQYGECL-
AAP13567.1 MYKTPTLKY-FGG-FNFSQILPDPLKPTKRSF---IEDLLFNKVTLADAGFMKQYGECL-
QHD43416.1 IYKTPPIKD-FGG-FNFSQILPDPSKPSKRSF---IEDLLFNKVTLADAGFIKQYGDCL-
AVP78031.1 IYKTPPIKD-FGG-FNFSQILPDPSKPSKRSF---IEDLLFNKVTLADAGFIKQYGDCL-
ABD75323.1 MYKTPTIRD-FGG-FNFSQILPDPLKPTKRSF---IEDLLYNKVTLADAGFMKQYADCL-
QDF43835.1 MYKTPAIKD-FGG-FNFSQILPDPSKPTKRSF---IEDLLFNKVTLADAGFMKQYGECL-
ABD75332.1 MYKTPAIKD-FGG-FNFSQILPDPSKPTKRSF---IEDLLFNKVTLADAGFMKQYGECL-
QDF43820.1 MYKTPAIKD-FGG-FNFSQILPDPSKPTKRSF---IEDLLFNKVTLADAGFMKQYGECL-
AAZ67052.1 MYKTPAIKD-FGG-FNFSQILPDPSKPTKRSF---IEDLLFNKVTLADAGFMKQYGECL-
AFS88936.1 SQSSPIIPG-FGGDFNLTLLEPVSISTGSRSARSAIEDLLFDKVTIADPGYMQGYDDCMQ
YP_0010399 T-STQTLEYGLNGDFNLTLLQVPQIGGSSSSYRSAIEDLLFDKVTIADPGYMQGYDDCMK
.: : :.* **:: : . * *****::***:**.*::: * :*:
QDF43825.1 -GDINARDLICAQKFNGLTVLPPLLTDDMIAAYTAALVSGTATAGWTFGAGAALQIPFAM
AGZ48818.1 -GDINARDLICAQKFNGLTVLPPLLTDDMIAAYTAALVSGTATAGWTFGAGAALQIPFAM
ALK02457.1 -GDINARDLICAQKFNGLTVLPPLLTDDMIAAYTAALVSGTATAGWTFGAGAALQIPFAM
AAS10463.1 -GDINARDLICAQKFNGLTVLPPLLTDDMIAAYTAALVSGTATAGWTFGAGAALQIPFAM
AAP13441.1 -GDINARDLICAQKFNGLTVLPPLLTDDMIAAYTAALVSGTATAGWTFGAGAALQIPFAM
AAP13567.1 -GDINARDLICAQKFNGLTVLPPLLTDDMIAAYTAALVSGTATAGWTFGAGAALQIPFAM
QHD43416.1 -GDIAARDLICAQKFNGLTVLPPLLTDEMIAQYTSALLAGTITSGWTFGAGAALQIPFAM
AVP78031.1 -GGISARDLICAQKFNGLTVLPPLLTDEMIAAYTAALISGTATAGWTFGAGAALQIPFAM
ABD75323.1 -GGINARDLICAQKFNGLTVLPPLLTDDMIAAYTAALISGTATAGWTFGAGAALQIPFAM
QDF43835.1 -GDINARDLICAQKFNGLTVLPPLLTDDMIAAYTAALVSGTATAGWTFGAGAALQIPFAM
ABD75332.1 -GDISARDLICAQKFNGLTVLPPLLTDEMIAAYTAALVSGTATAGWTFGAGSALQIPFAM
QDF43820.1 -GDINARDLICAQKFNGLTVLPPLLTDDMIAAYTAALVSGTATAGWTFGAGAALQIPFAM
AAZ67052.1 -GDISARDLICAQKFNGLTVLPPLLTDEMIAAYTAALVSGTATAGWTFGAGSALQIPFAM
AFS88936.1 QGPASARDLICAQYVAGYKVLPPLMDVNMEAAYTSSLLGSIAGVGWTAGLSSFAAIPFAQ
YP_0010399 QGPQSARDLICAQYVSGYKVLPPLYDPNMEAAYTSSLLGSIAGAGWTAGLSSFAAIPFAQ
* ******** . * .***** :* * **::*:.. *** * .: ****
QDF43825.1 QMAYRFNGIGVTQNVLYENQKQIANQFNKAISQIQESLTTTSTALGKLQDVVNQNAQALN
AGZ48818.1 QMAYRFNGIGVTQNVLYENQKQIANQFNKAISQIQESLTTTSTALGKLQDVVNQNAQALN
ALK02457.1 QMAYRFNGIGVTQNVLYENQKQIANQFNKAISQIQESLTTTSTALGKLQDVVNQNAQALN
AAS10463.1 QMAYRFNGIGVTQNVLYENQKQIANQFNKAISQIQESLTTTSTALGKLQDVVNQNAQALN
AAP13441.1 QMAYRFNGIGVTQNVLYENQKQIANQFNKAISQIQESLTTTSTALGKLQDVVNQNAQALN
AAP13567.1 QMAYRFNGIGVTQNVLYENQKQIANQFNKAISQIQESLTTTSTALGKLQDVVNQNAQALN
QHD43416.1 QMAYRFNGIGVTQNVLYENQKLIANQFNSAIGKIQDSLSSTASALGKLQDVVNQNAQALN
AVP78031.1 QMAYRFNGIGVTQNVLYENQKLIANQFNSAIGKIQESLTSTASALGKLQDVVNQNAQALN
ABD75323.1 QMAYRFNGIGVTQNVLYENQKQIANQFNKAITQIQESLTTTSTALGKLQDVVNQNAQALN
QDF43835.1 QMAYRFNGIGVTQNVLYENQKQIANQFNKAISQIQESLTTTSTALGKLQDVVNQNAQALN
ABD75332.1 QMAYRFNGIGVTQNVLYENQKQIANQFNKAISQIQESLTTTSTALGKLQDVVNQNAQALN
QDF43820.1 QMAYRFNGIGVTQNVLYENQKQIANQFNKAISQIQESLTTTSTALGKLQDVVNQNAQALN
AAZ67052.1 QMAYRFNGIGVTQNVLYENQKQIANQFNKAISQIQESLTTTSTALGKLQDVVNQNAQALN
AFS88936.1 SIFYRLNGVGITQQVLSENQKLIANKFNQALGAMQTGFTTTNEAFQKVQDAVNNNAQALS
YP_0010399 SMFYRLNGVGITQQVLSENQKLIANKFNQALGAMQTGFTTSNLAFSKVQDAVNANAQALS
.: **:**:*:**:** **** ***:**.*: :* .:::: *: *:**.** *****.
QDF43825.1 TLVKQLSSNFGAISSVLNDILSRLDKVEAEVQIDRLITGRLQSLQTYVTQQLIRAAEIRA
AGZ48818.1 TLVKQLSSNFGAISSVLNDILSRLDKVEAEVQIDRLITGRLQSLQTYVTQQLIRAAEIRA
ALK02457.1 TLVKQLSSNFGAISSVLNDILSRLDKVEAEVQIDRLITGRLQSLQTYVTQQLIRAAEIRA
AAS10463.1 TLVKQLSSNFGAISSVLNDILSRLDKVEAEVQIDRLITGRLQSLQTYVTQQLIRAAEIRA
AAP13441.1 TLVKQLSSNFGAISSVLNDILSRLDKVEAEVQIDRLITGRLQSLQTYVTQQLIRAAEIRA
AAP13567.1 TLVKQLSSNFGAISSVLNDILSRLDKVEAEVQIDRLITGRLQSLQTYVTQQLIRAAEIRA
QHD43416.1 TLVKQLSSNFGAISSVLNDILSRLDKVEAEVQIDRLITGRLQSLQTYVTQQLIRAAEIRA
AVP78031.1 TLVKQLSSNFGAISSVLNDILSRLDKVEAEVQIDRLITGRLQSLQTYVTQQLIRAAEIRA
ABD75323.1 TLVKQLSSNFGAISSALNDILSRLDKVEAEVQIDRLITGRLQSLQTYVTQQLIRAAEIRA
QDF43835.1 TLVKQLSSNFGAISSVLNDILSRLDKVEAEVQIDRLITGRLQSLQTYVTQQLIRAAEIRA
ABD75332.1 TLVKQLSSNFGAISSVLNDILSRLDKVEAEVQIDRLITGRLQSLQTYVTQQLIRAAEIRA
QDF43820.1 TLVKQLSSNFGAISSVLNDILSRLDKVEAEVQIDRLITGRLQSLQTYVTQQLIRAAEIRA
AAZ67052.1 TLVKQLSSNFGAISSVLNDILSRLDKVEAEVQIDRLITGRLQSLQTYVTQQLIRAAEIRA
AFS88936.1 KLASELSNTFGAISASIGDIIQRLDVLEQDAQIDRLINGRLTTLNAFVAQQLVRSESAAL
YP_0010399 KLASELSNTFGAISSSISDILARLDTVEQDAQIDRLINGRLISLNAFVSQQLVRSETAAR
.*..:**..*****: :.**: *** :* :.******.*** :*:::*:***:*:
QDF43825.1 SANLAATKMSECVLGQSKRVDFCGKGYHLMSFPQAAPHGVVFLHVTYVPSQERNFTTAPA
AGZ48818.1 SANLAATKMSECVLGQSKRVDFCGKGYHLMSFPQAAPHGVVFLHVTYVPSQERNFTTAPA
ALK02457.1 SANLAATKMSECVLGQSKRVDFCGKGYHLMSFPQAAPHGVVFLHVTYVPSQERNFTTAPA
AAS10463.1 SANLAATKMSECVLGQSKRVDFCGKGYHLMSFPQAAPHGVVFLHVTYVPSQERNFTTAPA
AAP13441.1 SANLAATKMSECVLGQSKRVDFCGKGYHLMSFPQAAPHGVVFLHVTYVPSQERNFTTAPA
AAP13567.1 SANLAATKMSECVLGQSKRVDFCGKGYHLMSFPQAAPHGVVFLHVTYVPSQERNFTTAPA
QHD43416.1 SANLAATKMSECVLGQSKRVDFCGKGYHLMSFPQSAPHGVVFLHVTYVPAQEKNFTTAPA
AVP78031.1 SANLAATKMSECVLGQSKRVDFCGKGYHLMSFPQSAPHGVVFLHVTYIPSQEKNFTTAPA
ABD75323.1 SANLAATKMSECVLGQSKRVDFCGKGYHLMSFPQSAPHGVVFLHVTYVPSQEKNFTTAPA
QDF43835.1 SANLAATKMSECVLGQSKRVDFCGRGYHLMSFPQAAPHGVVFLHVTYVPSQEKNFTTAPA
ABD75332.1 SANLAATKMSECVLGQSKRVDFCGKGYHLMSFPQAAPHGVVFLHVTYVPSQERNFTTAPA
QDF43820.1 SANLAATKMSECVLGQSKRVDFCGRGYHLMSFPQAAPHGVVFLHVTYVPSQEKNFTTAPA
AAZ67052.1 SANLAATKMSECVLGQSKRVDFCGKGYHLMSFPQAAPHGVVFLHVTYVPSQERNFTTAPA
AFS88936.1 SAQLAKDKVNECVKAQSKRSGFCGQGTHIVSFVVNAPNGLYFMHVGYYPSNHIEVVSAYG
YP_0010399 SAQLASDKVNECVKSQSKRNGFCGSGTHIVSFVVNAPNGFYFFHVGYVPTNYTNVTAAYG
**:** *:.*** .**** .*** * *::** **:*. *:** * *:: :..:* .
QDF43825.1 ICHEGK---AYFPREGVFVFNGTS-------WFITQRNFFSPQIITTDNT-FVSGSCDVV
AGZ48818.1 ICHEGK---AYFPREGVFVFNGTS-------WFITQRNFFSPQIITTDNT-FVSGSCDVV
ALK02457.1 ICHEGK---AYFPREGVFVFNGTS-------WFITQRNFFSPQIITTDNT-FVSGSCDVV
AAS10463.1 ICHEGK---AYFPREGVFVFNGTS-------WFITQRNFFSPQIITTDNT-FVSGNCDVV
AAP13441.1 ICHEGK---AYFPREGVFVFNGTS-------WFITQRNFFSPQIITTDNT-FVSGNCDVV
AAP13567.1 ICHEGK---AYFPREGVFVFNGTS-------WFITQRNFFSPQIITTDNT-FVSGNCDVV
QHD43416.1 ICHDGK---AHFPREGVFVSNGTH-------WFVTQRNFYEPQIITTDNT-FVSGNCDVV
AVP78031.1 ICHEGK---AHFPREGVFVSNGTH-------WFVTQRNFYEPKIITTDNT-FVSGNCDVV
ABD75323.1 ICHEGK---AYFPREGVFVSNGSS-------WFITQRNFYSPQIITTDNT-FVAGSCDVV
QDF43835.1 ICHEGK---AYFPREGVFVSNGTS-------WFITQRNFYSPQIITTDNT-FVAGSCDVV
ABD75332.1 ICHEGK---AYFPREGVFVSNGTS-------WFITQRNFYSPQIITTDNT-FVAGNCDVV
QDF43820.1 ICHEGK---AYFPREGVFVSNGTF-------WFITQRNFYSPQIITTDNT-FVAGNCDVV
AAZ67052.1 ICHEGK---AYFPREGVFVSNGTS-------WFITQRNFYSPQIITTDNT-FVAGSCDVV
AFS88936.1 LCDAANPTNCIAPVNGYFIKTNNT--RIVDEWSYTGSSFYAPEPITSLNTKYVA--PQVT
YP_0010399 LCNNNNPPLCIAPIDGYFITNQTTTYSVDTEWYYTGSSFYKPEPITQANSRYVS--SDVK
:* : . * :* *: . . * * .*: *: ** *: :*: :*
QDF43825.1 IGIINNTVYDPL---QPELDSFKEELDKYFKNHTSPDVDLGDISGINASVVNIQKEIDRL
AGZ48818.1 IGIINNTVYDPL---QPELDSFKEELDKYFKNHTSPDVDLGDISGINASVVNIQKEINRL
ALK02457.1 IGIINNTVYDPL---QPELDSFKEELDKYFKNHTSPDVDLGDISGINASVVNIQKEIDRL
AAS10463.1 IGIINNTVYDPL---QPELDSFKEELDKYFKNHTSPDVDLGDISGINASVVNIQEEIDRL
AAP13441.1 IGIINNTVYDPL---QPELDSFKEELDKYFKNHTSPDVDLGDISGINASVVNIQKEIDRL
AAP13567.1 IGIINNTVYDPL---QPELDSFKEELDKYFKNHTSPDVDLGDISGINASVVNIQKEIDRL
QHD43416.1 IGIVNNTVYDPL---QPELDSFKEELDKYFKNHTSPDVDLGDISGINASVVNIQKEIDRL
AVP78031.1 IGIINNTVYDPL---QPELDSFKEELDKYFKNHTSPDIDLGDISGINASVVNIQKEIDRL
ABD75323.1 IGIINNTVYDPL---QPELDSFKQELDKYFKNHTSPDVDLGDISGINASVVDIQKEIDRL
QDF43835.1 IGIINNTVYDPL---QPELDSFKEELDKYFKNHTSPDVDLGDISGINASVVNIQKEIDRL
ABD75332.1 IGIINNTVYDPL---QPELDSFKEELDKYFKNHTSPDVDLGDISGINASVVNIQKEIDRL
QDF43820.1 IGIINNTVYDPL---QPELDSFKEELDKYFKNHTSPDVDLGDISGINASVVNIQKEIDRL
AAZ67052.1 IGIINNTVYDPL---QPELDSFKEELDKYFKNHTSPDVDLGDISGINASVVNIQKEIDRL
AFS88936.1 YQNISTNLPPPLLGNSTGID-FQDELDEFFKNVSTSIPNFGSLTQINTTLLDLTYEMLSL
YP_0010399 FDKLENNLPPPLLENSTDVD-FKDELEEFFKNVTSHGPNFAEISKINTTLLDLSDEMAML
:...: ** .. :* *::**:::*** :: ::..:: **:::::: *: *
QDF43825.1 NEVAKNLNESLIDLQELGKYEQYIKWPWYVWLGFIAGLIAIVMVTILLCCMTSCCSCLKG
AGZ48818.1 NEVAKNLNESLIDLQELGKYEQYIKWPWYVWLGFIAGLIAIVMVTILLCCMTSCCSCLKG
ALK02457.1 NEVAKNLNESLIDLQELGKYEQYIKWPWYVWLGFIAGLIAIVMVTILLCCMTSCCSCLKG
AAS10463.1 NEVAKNLNESLIDLQELGKYEQYIKWPWYVWLGFIAGLIAIVMVTILLCCMTSCCSCLKG
AAP13441.1 NEVAKNLNESLIDLQELGKYEQYIKWPWYVWLGFIAGLIAIVMVTILLCCMTSCCSCLKG
AAP13567.1 NEVAKNLNESLIDLQELGKYEQYIKWPWYVWLGFIAGLIAIVMVTILLCCMTSCCSCLKG
QHD43416.1 NEVAKNLNESLIDLQELGKYEQYIKWPWYIWLGFIAGLIAIVMVTIMLCCMTSCCSCLKG
AVP78031.1 NEVARNLNESLIDLQELGKYEQYIKWPWYVWLGFIAGLIAIVMVTILLCCMTSCCSCLKG
ABD75323.1 NEVAKNLNESLIDLQELGKYEQYIKWPWYVWLGFIAGLVGLFMAIILLCYFTSCCSCCKG
QDF43835.1 NEVAKNLNESLIDLQELGKYEQYIKWPWYVWLGFIAGLIAIVMATILLCCMTSCCSCLKG
ABD75332.1 NEVAKNLNESLIDLQELGKYEQYIKWPWYVWLGFIAGLIAIVMVTILLCCMTSCCSCLKG
QDF43820.1 NEVAKNLNESLIDLQELGKYEQYIKWPWYVWLGFIAGLIAIVMATILLCCMTSCCSCLKG
AAZ67052.1 NEVAKNLNESLIDLQELGKYEQYIKWPWYVWLGFIAGLIAIVMVTILLCCMTSCCSCLKG
AFS88936.1 QQVVKALNESYIDLKELGNYTYYNKWPWYIWLGFIAGLVALALCVFFILCCTGCGTNCMG
YP_0010399 QEVVKQLNDSYIDLKELGNYTYYNKWPWYVWLGFIAGLVALLLCVFFLLCCTGCGTSCLG
::*.. **:* ***:***:* * *****:********:.: : ::: *.* : *
QDF43825.1 ACSCGSCC-KFDEDDSEPVLKGVKLHYT
AGZ48818.1 ACSCGSCC-KFDEDDSEPVLKGVKLHYT
ALK02457.1 ACSCGSCC-KFDEDDSEPVLKGVKLHYT
AAS10463.1 ACSCGSCC-KFDEDDSEPVLKGVKLHYT
AAP13441.1 ACSCGSCC-KFDEDDSEPVLKGVKLHYT
AAP13567.1 ACSCGSCC-KFDEDDSEPVLKGVKLHYT
QHD43416.1 CCSCGSCC-KFDEDDSEPVLKGVKLHYT
AVP78031.1 CCSCGSCC-KFDEDDSEPVLKGVKLHYT
ABD75323.1 MCSCGSCC-RFDEDDSEPVLKGVKLHYT
QDF43835.1 ACSCGSCC-KFDEDDSEPVLKGVKLHYT
ABD75332.1 ACSCGSCC-KFDEDDSEPVLKGVKLHYT
QDF43820.1 ACSCGSCC-KFDEDDSEPVLKGVKLHYT
AAZ67052.1 ACSCGSCC-KFDEDDSEPVLKGVKLHYT
AFS88936.1 KLKCNRCCDRYEEYDLEP----HKVHVH
YP_0010399 KMKCKNCCDSYEEYDVE------KIHVH
.* ** ::* * * *:*
Phylogenetic Tree
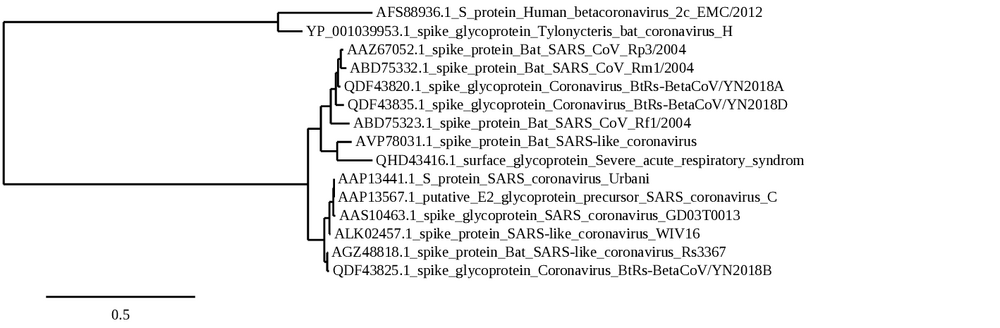
Figure 1: Phylogenetic tree of 15 beta coronavirus spike protein sequences. 0.5 scale line is provided for comparative purposes. A horizontal distance of 0.5 denotes 50% evolutionary divergence. Vertical distances do not denote divergence.
Receptor Binding Domain Sequence
The sequences provided below are the residues of the spike protein in the Receptor Binding Domain. Residues included in the RBD are marked to start at the marker | and end at the marker !.
START |
QDF43825.1 AVDCSQNPLAELKCSVKSFEIDKGIYQTSNFRVAPSKEVVRFPNITNLCPFGEVFNATTF
AGZ48818.1 AVDCSQNPLAELKCSVKSFEIDKGIYQTSNFRVAPSKEVVRFPNITNLCPFGEVFNATTF
ALK02457.1 AVDCSQNPLAELKCSVKSFEIDKGIYQTSNFRVAPSKEVVRFPNITNLCPFGEVFNATTF
AAS10463.1 AVDCSQNPLAELKCSVKSFEIDKGIYQTSNFRVVPSGDVVRFPNITNLCPFGEVFNATKF
AAP13441.1 AVDCSQNPLAELKCSVKSFEIDKGIYQTSNFRVVPSGDVVRFPNITNLCPFGEVFNATKF
AAP13567.1 AVDCSQNPLAELKCSVKSFEIDKGIYQTSNFRVVPSGDVVRFPNITNLCPFGEVFNATKF
QHD43416.1 AVDCALDPLSETKCTLKSFTVEKGIYQTSNFRVQPTESIVRFPNITNLCPFGEVFNATRF
AVP78031.1 AVDCALDPLSETKCTLKSLTVQKGIYQTSNFRVQPTQSVVRFPNITNVCPFHKVFNATRF
ABD75323.1 AVDCSQDPLAELKCTTKSFNVSKGIYQTSNFRVSPVTEVVRFPNITNLCPFDKVFNATRF
QDF43835.1 AIDCAQNPLSELKCTIKNFNVSKGIYQTSNFRVSPTHEVIRFPNITNRCPFDKVFNASRF
ABD75332.1 AVDCSQNPLAELKCTIKNFNVSKGIYQTSNFRVTPTQEVVRFPNITNRCPFDKVFNASRF
QDF43820.1 AIDCAQNPLAELKCTIKNFNVSKGIYQTSNFRVSPTQEVVRFPNITNRCPFDKVFNASRF
AAZ67052.1 AIDCAQNPLAELKCTIKNFNVSKGIYQTSNFRVSPTQEVIRFPNITNRCPFDKVFNATRF
AFS88936.1 AIDCGFNDLSQLHCSYESFDVESGVYSVSSFEAKPSGSVVEQAEGVE-CDFSPLLSGTP-
YP_0010399 AIDCGHDDLSQLHCSYTSFEVDTGVYSVSSYEASATGTFIEQPNATE-CDFSPMLTGVA-
*:**. : *:: :*: .: :..*:*..*.: . . .: .: .: * * ::..
QDF43825.1 PSVYAWERKRISNCVADYSVLYNSTSFSTFKCYGVSATKLNDLCFSNVYADSFVVKGDDV
AGZ48818.1 PSVYAWERKRISNCVADYSVLYNSTSFSTFKCYGVSATKLNDLCFSNVYADSFVVKGDDV
ALK02457.1 PSVYAWERKRISNCVADYSVLYNSTSFSTFKCYGVSATKLNDLCFSNVYADSFVVKGDDV
AAS10463.1 PSVYAWERKRISNCVADYSVLYNSTSFSTFKCYGVSATKLNDLCFSNVYADSFVVKGDDV
AAP13441.1 PSVYAWERKKISNCVADYSVLYNSTFFSTFKCYGVSATKLNDLCFSNVYADSFVVKGDDV
AAP13567.1 PSVYAWERKKISNCVADYSVLYNSTFFSTFKCYGVSATKLNDLCFSNVYADSFVVKGDDV
QHD43416.1 ASVYAWNRKRISNCVADYSVLYNSASFSTFKCYGVSPTKLNDLCFTNVYADSFVIRGDEV
AVP78031.1 PSVYAWERTKISDCIADYTVFYNSTSFSTFKCYGVSPSKLIDLCFTSVYADTFLIRFSEV
ABD75323.1 PSVYAWERTKISDCVADYTVFYNSTSFSTFNCYGVSPSKLIDLCFTSVYADTFLIRFSEV
QDF43835.1 PNVYAWERTKISDCVADYTVLYNSTSFSTFKCYGVSPSKLIDLCFTSVYADTFLIRSSEV
ABD75332.1 PNVYAWERTKISDCVADYTVLYNSTSFSTFKCYGVSPSKLIDLCFTSVYADTFLIRSSEV
QDF43820.1 PNVYAWERTKISDCVADYTVLYNSTSFSTFKCYGVSPSKLIDLCFTSVYADTFLIRSSEV
AAZ67052.1 PNVYAWERTKISDCVADYTVLYNSTSFSTFKCYGVSPSKLIDLCFTSVYADTFLIRSSEV
AFS88936.1 PQVYNFKRLVFTNCNYNLTKLLSLFSVNDFTCSQISPAAIASNCYSSLILDYFSYPLSMK
YP_0010399 PQVYNFKRLVFSNCNYNLTKLLSLFAVDEFSCNGISPDSIARGCYSTLTVDYFAYPLSMK
..** ::* :::* : : : . .. *.* :*. : *::.: * * .
QDF43825.1 RQIAPGQTGVIADYNYKLPDDFMGC-VLAWNTRNIDATSTGNYNYKYRSLRHGKLRPFER
AGZ48818.1 RQIAPGQTGVIADYNYKLPDDFTGC-VLAWNTRNIDATQTGNYNYKYRSLRHGKLRPFER
ALK02457.1 RQIAPGQTGVIADYNYKLPDDFTGC-VLAWNTRNIDATQTGNYNYKYRSLRHGKLRPFER
AAS10463.1 RQIAPGQTGVIADYNYKLPDDFMGC-VLAWNTRNIDATSTGNYNYKYRYLRHGKLRPFER
AAP13441.1 RQIAPGQTGVIADYNYKLPDDFMGC-VLAWNTRNIDATSTGNYNYKYRYLRHGKLRPFER
AAP13567.1 RQIAPGQTGVIADYNYKLPDDFMGC-VLAWNTRNIDATSTGNYNYKYRYLRHGKLRPFER
QHD43416.1 RQIAPGQTGKIADYNYKLPDDFTGC-VIAWNSNNLDSKVGGNYNYLYRLFRKSNLKPFER
AVP78031.1 RQVAPGQTGVIADYNYKLPDDFTGC-VIAWNTAKQD---VGNYF--YRSHRSTKLKPFER
ABD75323.1 RQVAPGQTGVIADYNYKLPDDFTGC-VIAWNTAKQD---VGSYF--YRSHRSSKLKPFER
QDF43835.1 RQVAPGETGVIADYNYKLPDDFTGC-VIAWNTAKQD---QGQYY--YRSSRKTKLKPFER
ABD75332.1 RQVAPGETGVIADYNYKLPDDFTGC-VIAWNTAQQD---QGQYY--YRSYRKEKLKPFER
QDF43820.1 RQVAPGETGVIADYNYKLPDDFTGC-VIAWNTAKQD---TGHYY--YRSHRKTKLKPFER
AAZ67052.1 RQVAPGETGVIADYNYKLPDDFTGC-VIAWNTAKQD---QGQYY--YRSHRKTKLKPFER
AFS88936.1 SDLSVSSAGPISQFNYKQSFSNPTC-LILATVPHNLTTITKPLKYSYINKCSRLLSDDRT
YP_0010399 SYIRPGSAGNIPLYNYKQSFANPTCRVMASVLANVTITKPHAYG--YIS-KCSRLTGANQ
: ..:* *. :*** . * :: : * *
QDF43825.1 DISNVPFSPDGKPCTPP-AF-NCYW-----------PLNDYGFFTTNGIGYQPYRVVVLS
AGZ48818.1 DISNVPFSPDGKPCTPP-AF-NCYW-----------PLNDYGFYITNGIGYQPYRVVVLS
ALK02457.1 DISNVPFSPDGKPCTPP-AF-NCYW-----------PLNDYGFYITNGIGYQPYRVVVLS
AAS10463.1 DISNVPFSPDGKPCTPP-AP-NCYW-----------PLNGYGFYTTSGIGYQPYRVVVLS
AAP13441.1 DISNVPFSPDGKPCTPP-AL-NCYW-----------PLNDYGFYTTTGIGYQPYRVVVLS
AAP13567.1 DISNVPFSPDGKPCTPP-AL-NCYW-----------PLNDYGFYTTTGIGYQPYRVVVLS
QHD43416.1 DISTEIYQAGSTPCNGVEGF-NCYF-----------PLQSYGFQPTNGVGYQPYRVVVLS
AVP78031.1 DLSSDE---------------NGVR-----------TLSTYDFNPNVPLEYQATRVVVLS
ABD75323.1 DLSSEE---------------NGVR-----------TLSTYDFNQNVPLEYQATRVVVLS
QDF43835.1 DLTSDE---------------NGVR-----------TLSTYDFYPNVPIEYQATRVVVLS
ABD75332.1 DLSSDE---------------NGVY-----------TLSTYDFYPSIPVEYQATRVVVLS
QDF43820.1 DLSSDDG--------------NGVY-----------TLSTYDFNPNVPVAYQATRVVVLS
AAZ67052.1 DLSSDE---------------NGVR-----------TLSTYDFYPSVPVAYQATRVVVLS
AFS88936.1 EVPQLVNANQYSPCVSI-VP-STVWEDGDYYRKQLSPLEGGGWLVASGSTVAMTEQLQMG
YP_0010399 DVETPLYINPGEYSICRDFSPGGFSEDGQVFKRTLTQFEGGGLLIGVGTRVPMTDNLQMS
:: . :. . : :.
QDF43825.1 FELL----NAPATVC-----GPKLSTDLIKNQCVNFNFNGLTGTGVLTPSSKRFQPFQQF
AGZ48818.1 FELL----NAPATVC-----GPKLSTDLIKNQCVNFNFNGLTGTGVLTPSSKRFQPFQQF
ALK02457.1 FELL----NAPATVC-----GPKLSTDLIKNQCVNFNFNGLTGTGVLTPSSKRFQPFQQF
AAS10463.1 FELL----NAPATVC-----GPKLSTDLIKNQCVNFNFNGLTGTGVLTPSSKRFQPFQQF
AAP13441.1 FELL----NAPATVC-----GPKLSTDLIKNQCVNFNFNGLTGTGVLTPSSKRFQPFQQF
AAP13567.1 FELL----NAPATVC-----GPKLSTDLIKNQCVNFNFNGLTGTGVLTPSSKRFQPFQQF
QHD43416.1 FELL----HAPATVC-----GPKKSTNLVKNKCVNFNFNGLTGTGVLTESNKKFLPFQQF
AVP78031.1 FELL----NAPATVC-----GPKLSTQLVKNQCVNFNFNGLKGTGVLTDSSKRFQSFQQF
ABD75323.1 FELL----NAPATVC-----GPKLSTSLVKNQCVNFNFNGFKGTGVLTDSSKTFQSFQQF
QDF43835.1 FELL----NAPATVC-----GPKLSTGLVKNQCVNFNFNGLRGTGVLTDSSKRFQSFQQF
ABD75332.1 FELL----NAPATVC-----GPKLSTQLVKNQCVNFNFNGLRGTGVLTTSSKRFQSFQQF
QDF43820.1 FELL----NAPATVC-----GPKLSTQLVKNQCVNFNFNGLKGTGVLTDSSKRFQSFQQF
AAZ67052.1 FELL----NAPATVC-----GPKLSTQLVKNQCVNFNFNGLKGTGVLTESSKRFQSFQQF
AFS88936.1 FGITVQYGTDTNSVCPKLEFANDTKIASQLGNCVEYSLYGVSGRGVFQNCTAVGVRQQRF
YP_0010399 FIISVQYGTGTDSVCPMLDLGDSLTITNRLGKCVDYSLYGVTGRGVFQNCTAVGVKQQRF
* : . :** . . . .:**::.: *. * **: .. *.*
END !
Scientific Conclusion
A phylogenetic tree was created using the sequences of 15 spike proteins from beta-coronaviruses. The spike protein sequences were then aligned and assessed for divergence including a comparison of alignment with the phylogram. Both phylogram and sequence alignment were compared with those provided in Wan et al 2020. Ultimately, this study determined that SARS-CoV-2 spike proteins are highly evolutionarily divergent from related beta-coronaviruses, including SARS-CoV-1.
Acknowledgements
- Protocol was copied from [BIOL368/F20:Week_4] and edited to fit exact methods used
- Collaboration took place with Owen Daily on successful completion of phylogenetic tree and concerning the differences of the SARS-CoV-2 spike protein sequence to the other beta-coronaviruses assessed.
- Dr. Kam D. Dahlquist for tutorials on phylogeny.fr and how to interpret phylogenetic trees.
- Except for what is noted above, this individual journal entry was completed by me and not copied from another source.
References
- Dereeper A., Audic S., Claverie J.M., Blanc G. BLAST-EXPLORER helps you building datasets for phylogenetic analysis. BMC Evol Biol. 2010 Jan 12;10:8. (PubMed)
- Dereeper A.*, Guignon V.*, Blanc G., Audic S., Buffet S., Chevenet F., Dufayard J.F., Guindon S., Lefort V., Lescot M., Claverie J.M., Gascuel O. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008 Jul 1;36(Web Server issue):W465-9. Epub 2008 Apr 19. (PubMed) *: joint first authors
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, Mar 19;32(5):1792-7. (PubMed)
- Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000, Apr;17(4):540-52. (PubMed)
- Guindon S., Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003, Oct;52(5):696-704. (PubMed)
- Anisimova M., Gascuel O. Approximate likelihood ratio test for branchs: A fast, accurate and powerful alternative. Syst Biol. 2006, Aug;55(4):539-52. (PubMed)
- Wan, Y., Shang, J., Graham, R., Baric, R. S., & Li, F. (2020). Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. Journal of virology, 94(7).