BIOL368/S19 Week 4
- Weekly Assignments
- Individual Journal Entries
- Fatimah Alghanem Journal Entry Week 2
- Fatimah Alghanem Journal Entry Week 3
- Fatimah Alghanem Journal Entry Week 4
- Fatimah Alghanem Journal Entry Week 5
- Fatimah Alghanem Journal Entry Week 6
- Fatimah Alghanem Journal Entry Week 7
- DrugComboDB Review
- Fatimah Alghanem Journal Entry Week 9
- Fatimah Alghanem Journal Entry Week 10
- Fatimah Alghanem Journal Entry Week 11
- The Mutants Research Project Week 12
- Fatimah Alghanem Journal Entry Week 14
- Class Journal
BIOL368/F20 BIOL368/F20:People
Purpose
The purpose of this assignment is to use GenBank to find genetic sequences. Moreover to learn how to find protein sequence and compare sequences with one another and to be able to create phylogenetic trees and compare them too.
Methods and results
Part 1: GneBank
- Coronavirus BtRs-BetaCoV/YN2018B GenBank records were chosen and both the full record and the FASTA formatted sequence were viewed.
- An accession number of the sequence was chosen: MK211376
- Information provided in the GenBank record was noted.
- The nucleotide sequence was downloaded in FASTA formate to my local hard drive
- The file was then opened in Word processor to make sure that it was downloaded and is in FASTA format, in FASTA format each sequence is preceded by a label which begins with the greater than sign (>).
- Links have been provided to the individual spike protein sequences corresponding to each of the viral genome records listed in the Data & Tools section.
- I was assigned a nucleotide sequence accession number from Figure 2 in class.
- spike glycoprotein [SARS coronavirus GD03T0013
- The GenBank record associated with that sequence was searched and a hyperlink to the GenBank record was added to the list of sequences in the Data & Tools section.
- The spike protein accession number in the GenBank was recorded: AAS10463
- A hyperlink to the spike protein record to the list of sequences in the Data & Tools section was added with making sure that to format the list in the same way as it is already formatted.
- Assigned protein sequence was downloaded in FASTA format like it was done for the whole genome sequence.
- The protein sequence was added to the "Talk" page.
- I was assigned a nucleotide sequence accession number from Figure 2 in class.
>AAS10463.1 spike glycoprotein [SARS coronavirus
GD03T0013]MFIFLLFLTLTSGSDLDRCTTFDDVQAPNYTQHTSSMRGVYYPDEIFRSDTLYLTQDLFLPFYSNVTGFHTINHTFDDPVIPFK DGIYFAATEKSNVVRGWVFGSTMNNKSQSVIIINNSTNVVIRACNFELCDNPFFVVSKPMGTRTHTMIFDNAFNCTFEYISDAFSLDVSEKS GNFKHLREFVFKNKDGFLYVYKGYQPIDVVRDLPSGFNTLKPIFKLPLGINITNFRAILTAFSPAQDTWGTSAAAYFVGYLKPTTFMLKYDEN GTITDAVDCSQNPLAELKCSVKSFEIDKGIYQTSNFRVVPSGDVVRFPNITNLCPFGEVFNATKFPSVYAWERKRISNCVADYSVLYNSTSFST FKCYGVSATKLNDLCFSNVYADSFVVKGDDVRQIAPGQTGVIADYNYKLPDDFMGCVLAWNTRNIDATSTGNYNYKYRYLRHGKLRPFER DISNVPFSPDGKPCTPPAPNCYWPLNGYGFYTTSGIGYQPYRVVVLSFELLNAPATVCGPKLSTDLIKNQCVNFNFNGLTGTGVLTPSSKRF QPFQQFGRDVSDFTDSVRDPKTSEILDISPCSFGGVSVITPGTNASSEVAVLYQDVNCTDVSTLIHAEQLTPAWRIYSTGNNVFQTQAGCLI. GAEHVDTSYECDIPIGAGICASYHTVSSLRSTSQKSIVAYTMSLGADSSIAYSNNTIAIPTNFSISITTEVMPVSMAKTSVDCNMYICGDSTECA NLLLQYGSFCRQLNRALSGIAAEQDRNTREVFVQVKQMYKTPTLKDFGGFNFSQILPDPLKPTKRSFIEDLLFNKVTLADAGFMKQYGECLG DINARDLICAQKFNGLTVLPPLLTDDMIAAYTAALVSGTATAGWTFGAGAALQIPFAMQMAYRFNGIGVTQNVLYENQKQIANQFNKAIS QIQESLTTTSTALGKLQDVVNQNAQALNTLVKQLSSNFGAISSVLNDILSRLDKVEAEVQIDRLITGRLQSLQTYVTQQLIRAAEIRASANLAA TKMSECVLGQSKRVDFCGKGYHLMSFPQAAPHGVVFLHVTYVPSQERNFTTAPAICHEGKAYFPREGVFVFNGTSWFITQRNFFSPQIITT DNTFVSGNCDVVIGIINNTVYDPLQPELDSFKEELDKYFKNHTSPDVDLGDISGINASVVNIQEEIDRLNEVAKNLNESLIDLQELGKYEQYIK WPWYVWLGFIAGLIAIVMVTILLCCMTSCCSCLKGACSCGSCCKFDEDDSEPVLKGVKLHYT
figure 1. protein sequance for AAS10463
Part 2: Creating a phylogenetic tree with Phylogeny.fr
- the Phylogeny.fr, a free, simple to use web service dedicated to reconstructing and analyzing phylogenetic relationships between molecular sequences was used to analyze sequence data.
- In the browser, the website www.phylogeny.fr. was opened, down on the page there was a section labeled ‘Phylogeny analysis’, and it was clicked on the text ‘One Click’.
- Then it was Clicked in the large text field labeled ‘Upload your set of sequences in FASTA, EMBL, or NEXUS format’. Then the list of sequences from the talk page was Copied and Command-V was used to paste the sequences there, then the “Submit” button was clicked.
- a page named Alignment results was seen. After alignment was complete, a new page named Phylogeny results was seen. Finally, a page named Tree rendering results was seen. Those pages were used later in the methods. For this part of the methods, the numbered tabs located just beneath the text One Click Mode was found, and the tab labeled 3. The alignment was clicked. Individual positions were color-coded to indicate their conservation, or how similar the sequences are to each other. Blue highlights indicated high conservation, while gray highlights indicated lower conservation and white highlights indicated little if any conservation.
- Near the bottom of the page, under Outputs, Alignment in Clustal format was clicked. This displayed alignment in a text-only format in which each position's conservation was indicated by a symbol underneath the alignment block (“*” for invariant, “:” for highly conserved, “.” for weakly conserved, and a space for not conserved). The entire alignment was then copied and pasted into the individual journal entry and the space character at the beginning of each line was used so that the sequence lines up properly on the page.
>QHD43416.1 surface glycoprotein [Severe acute respiratory syndrome coronavirus 2] MFVFLVLLPLVSSQCVNLTTRTQLPPAYTNSFTRGVYYPDKVFRSSVLHSTQDLFLPFFSNVTWFHAIHV SGTNGTKRFDNPVLPFNDGVYFASTEKSNIIRGWIFGTTLDSKTQSLLIVNNATNVVIKVCEFQFCNDPF LGVYYHKNNKSWMESEFRVYSSANNCTFEYVSQPFLMDLEGKQGNFKNLREFVFKNIDGYFKIYSKHTPI NLVRDLPQGFSALEPLVDLPIGINITRFQTLLALHRSYLTPGDSSSGWTAGAAAYYVGYLQPRTFLLKYN ENGTITDAVDCALDPLSETKCTLKSFTVEKGIYQTSNFRVQPTESIVRFPNITNLCPFGEVFNATRFASV YAWNRKRISNCVADYSVLYNSASFSTFKCYGVSPTKLNDLCFTNVYADSFVIRGDEVRQIAPGQTGKIAD YNYKLPDDFTGCVIAWNSNNLDSKVGGNYNYLYRLFRKSNLKPFERDISTEIYQAGSTPCNGVEGFNCYF PLQSYGFQPTNGVGYQPYRVVVLSFELLHAPATVCGPKKSTNLVKNKCVNFNFNGLTGTGVLTESNKKFL PFQQFGRDIADTTDAVRDPQTLEILDITPCSFGGVSVITPGTNTSNQVAVLYQDVNCTEVPVAIHADQLT PTWRVYSTGSNVFQTRAGCLIGAEHVNNSYECDIPIGAGICASYQTQTNSPRRARSVASQSIIAYTMSLG AENSVAYSNNSIAIPTNFTISVTTEILPVSMTKTSVDCTMYICGDSTECSNLLLQYGSFCTQLNRALTGI AVEQDKNTQEVFAQVKQIYKTPPIKDFGGFNFSQILPDPSKPSKRSFIEDLLFNKVTLADAGFIKQYGDC LGDIAARDLICAQKFNGLTVLPPLLTDEMIAQYTSALLAGTITSGWTFGAGAALQIPFAMQMAYRFNGIG VTQNVLYENQKLIANQFNSAIGKIQDSLSSTASALGKLQDVVNQNAQALNTLVKQLSSNFGAISSVLNDI LSRLDKVEAEVQIDRLITGRLQSLQTYVTQQLIRAAEIRASANLAATKMSECVLGQSKRVDFCGKGYHLM SFPQSAPHGVVFLHVTYVPAQEKNFTTAPAICHDGKAHFPREGVFVSNGTHWFVTQRNFYEPQIITTDNT FVSGNCDVVIGIVNNTVYDPLQPELDSFKEELDKYFKNHTSPDVDLGDISGINASVVNIQKEIDRLNEVA KNLNESLIDLQELGKYEQYIKWPWYIWLGFIAGLIAIVMVTIMLCCMTSCCSCLKGCCSCGSCCKFDEDD SEPVLKGVKLHYT
>AAP13441.1 S protein [SARS coronavirus Urbani] MFIFLLFLTLTSGSDLDRCTTFDDVQAPNYTQHTSSMRGVYYPDEIFRSDTLYLTQDLFLPFYSNVTGFH TINHTFGNPVIPFKDGIYFAATEKSNVVRGWVFGSTMNNKSQSVIIINNSTNVVIRACNFELCDNPFFAV SKPMGTQTHTMIFDNAFNCTFEYISDAFSLDVSEKSGNFKHLREFVFKNKDGFLYVYKGYQPIDVVRDLP SGFNTLKPIFKLPLGINITNFRAILTAFSPAQDIWGTSAAAYFVGYLKPTTFMLKYDENGTITDAVDCSQ NPLAELKCSVKSFEIDKGIYQTSNFRVVPSGDVVRFPNITNLCPFGEVFNATKFPSVYAWERKKISNCVA DYSVLYNSTFFSTFKCYGVSATKLNDLCFSNVYADSFVVKGDDVRQIAPGQTGVIADYNYKLPDDFMGCV LAWNTRNIDATSTGNYNYKYRYLRHGKLRPFERDISNVPFSPDGKPCTPPALNCYWPLNDYGFYTTTGIG YQPYRVVVLSFELLNAPATVCGPKLSTDLIKNQCVNFNFNGLTGTGVLTPSSKRFQPFQQFGRDVSDFTD SVRDPKTSEILDISPCSFGGVSVITPGTNASSEVAVLYQDVNCTDVSTAIHADQLTPAWRIYSTGNNVFQ TQAGCLIGAEHVDTSYECDIPIGAGICASYHTVSLLRSTSQKSIVAYTMSLGADSSIAYSNNTIAIPTNF SISITTEVMPVSMAKTSVDCNMYICGDSTECANLLLQYGSFCTQLNRALSGIAAEQDRNTREVFAQVKQM YKTPTLKYFGGFNFSQILPDPLKPTKRSFIEDLLFNKVTLADAGFMKQYGECLGDINARDLICAQKFNGL TVLPPLLTDDMIAAYTAALVSGTATAGWTFGAGAALQIPFAMQMAYRFNGIGVTQNVLYENQKQIANQFN KAISQIQESLTTTSTALGKLQDVVNQNAQALNTLVKQLSSNFGAISSVLNDILSRLDKVEAEVQIDRLIT GRLQSLQTYVTQQLIRAAEIRASANLAATKMSECVLGQSKRVDFCGKGYHLMSFPQAAPHGVVFLHVTYV PSQERNFTTAPAICHEGKAYFPREGVFVFNGTSWFITQRNFFSPQIITTDNTFVSGNCDVVIGIINNTVY DPLQPELDSFKEELDKYFKNHTSPDVDLGDISGINASVVNIQKEIDRLNEVAKNLNESLIDLQELGKYEQ YIKWPWYVWLGFIAGLIAIVMVTILLCCMTSCCSCLKGACSCGSCCKFDEDDSEPVLKGVKLHYT
>QDF43825.1 spike glycoprotein [Coronavirus BtRs-BetaCoV/YN2018B] MKLLVLVFATLVSSYTIEKCTDFDDRTPPSNTQFLSSHRGVYYPDDIFRSNVLHLVQDHFLPFDSNVTRF ITFGLNFDNPIIPFRDGVYFAATEKSNVIRGWVFGSTMNNKSQSVIIMNNSTNLVIRACNFELCDNPFFV VLRSNNTQIPSYIFNNAFNCTFEYVSKDFNLDIGEKPGNFKDLREFVFRNKDGFLHVYSGYQPISAASGL PTGFNALKPIFKLPLGINITNFRTLLTAFPPNPGYWGTSAAAYFVGYLKPTTFMLKYDENGTITDAVDCS QNPLAELKCSVKSFEIDKGIYQTSNFRVAPSKEVVRFPNITNLCPFGEVFNATTFPSVYAWERKRISNCV ADYSVLYNSTSFSTFKCYGVSATKLNDLCFSNVYADSFVVKGDDVRQIAPGQTGVIADYNYKLPDDFMGC VLAWNTRNIDATSTGNYNYKYRSLRHGKLRPFERDISNVPFSPDGKPCTPPAFNCYWPLNDYGFFTTNGI GYQPYRVVVLSFELLNAPATVCGPKLSTDLIKNQCVNFNFNGLTGTGVLTPSSKRFQPFQQFGRDVSDFT DSVRDPKTSEILDISPCSFGGVSVITPGTNTSSEVAVLYQDVNCTDVPVAIHADQLTPAWRIYSTGNNVF QTQAGCLIGAEHVDTSYECDIPIGAGICASYHTVSSLRSTSQKSIVAYTMSLGADSSIAYSNNTIAIPTN FSISITTEVMPVSMAKTSVDCNMYICGDSTECANLLLQYGSFCTQLNRALSGIAVEQDRNTREVFAQVKQ MYKTPTLKDFGGFNFSQILPDPLKPTKRSFIEDLLFNKVTLADAGFMKQYGECLGDINARDLICAQKFNG LTVLPPLLTDDMIAAYTAALVSGTATAGWTFGAGAALQIPFAMQMAYRFNGIGVTQNVLYENQKQIANQF NKAISQIQESLTTTSTALGKLQDVVNQNAQALNTLVKQLSSNFGAISSVLNDILSRLDKVEAEVQIDRLI TGRLQSLQTYVTQQLIRAAEIRASANLAATKMSECVLGQSKRVDFCGKGYHLMSFPQAAPHGVVFLHVTY VPSQERNFTTAPAICHEGKAYFPREGVFVFNGTSWFITQRNFFSPQIITTDNTFVSGSCDVVIGIINNTV YDPLQPELDSFKEELDKYFKNHTSPDVDLGDISGINASVVNIQKEIDRLNEVAKNLNESLIDLQELGKYE QYIKWPWYVWLGFIAGLIAIVMVTILLCCMTSCCSCLKGACSCGSCCKFDEDDSEPVLKGVKLHYT
>AFS88936.1 S protein [Human betacoronavirus 2c EMC/2012] MIHSVFLLMFLLTPTESYVDVGPDSVKSACIEVDIQQTFFDKTWPRPIDVSKADGIIYPQGRTYSNITIT YQGLFPYQGDHGDMYVYSAGHATGTTPQKLFVANYSQDVKQFANGFVVRIGAAANSTGTVIISPSTSATI RKIYPAFMLGSSVGNFSDGKMGRFFNHTLVLLPDGCGTLLRAFYCILEPRSGNHCPAGNSYTSFATYHTP ATDCSDGNYNRNASLNSFKEYFNLRNCTFMYTYNITEDEILEWFGITQTAQGVHLFSSRYVDLYGGNMFQ FATLPVYDTIKYYSIIPHSIRSIQSDRKAWAAFYVYKLQPLTFLLDFSVDGYIRRAIDCGFNDLSQLHCS YESFDVESGVYSVSSFEAKPSGSVVEQAEGVECDFSPLLSGTPPQVYNFKRLVFTNCNYNLTKLLSLFSV NDFTCSQISPAAIASNCYSSLILDYFSYPLSMKSDLSVSSAGPISQFNYKQSFSNPTCLILATVPHNLTT ITKPLKYSYINKCSRLLSDDRTEVPQLVNANQYSPCVSIVPSTVWEDGDYYRKQLSPLEGGGWLVASGST VAMTEQLQMGFGITVQYGTDTNSVCPKLEFANDTKIASQLGNCVEYSLYGVSGRGVFQNCTAVGVRQQRF VYDAYQNLVGYYSDDGNYYCLRACVSVPVSVIYDKETKTHATLFGSVACEHISSTMSQYSRSTRSMLKRR DSTYGPLQTPVGCVLGLVNSSLFVEDCKLPLGQSLCALPDTPSTLTPRSVRSVPGEMRLASIAFNHPIQV DQLNSSYFKLSIPTNFSFGVTQEYIQTTIQKVTVDCKQYVCNGFQKCEQLLREYGQFCSKINQALHGANL RQDDSVRNLFASVKSSQSSPIIPGFGGDFNLTLLEPVSISTGSRSARSAIEDLLFDKVTIADPGYMQGYD DCMQQGPASARDLICAQYVAGYKVLPPLMDVNMEAAYTSSLLGSIAGVGWTAGLSSFAAIPFAQSIFYRL NGVGITQQVLSENQKLIANKFNQALGAMQTGFTTTNEAFQKVQDAVNNNAQALSKLASELSNTFGAISAS IGDIIQRLDVLEQDAQIDRLINGRLTTLNAFVAQQLVRSESAALSAQLAKDKVNECVKAQSKRSGFCGQG THIVSFVVNAPNGLYFMHVGYYPSNHIEVVSAYGLCDAANPTNCIAPVNGYFIKTNNTRIVDEWSYTGSS FYAPEPITSLNTKYVAPQVTYQNISTNLPPPLLGNSTGIDFQDELDEFFKNVSTSIPNFGSLTQINTTLL DLTYEMLSLQQVVKALNESYIDLKELGNYTYYNKWPWYIWLGFIAGLVALALCVFFILCCTGCGTNCMGK LKCNRCCDRYEEYDLEPHKVHVH
>YP_001039953.1 spike glycoprotein [Tylonycteris bat coronavirus HKU4] MTLLMCLLMSLLIFVRGCDSQFVDMSPASNTSECLESQVDAAAFSKLMWPYPIDPSKVDGIIYPLGRTYS NITLAYTGLFPLQGDLGSQYLYSVSHAVGHDGDPTKAYISNYSLLVNDFDNGFVVRIGAAANSTGTIVIS PSVNTKIKKAYPAFILGSSLTNTSAGQPLYANYSLTIIPDGCGTVLHAFYCILKPRTVNRCPSGTGYVSY FIYETVHNDCQSTINRNASLNSFKSFFDLVNCTFFNSWDITADETKEWFGITQDTQGVHLYSSRKGDLYG GNMFRFATLPVYEGIKYYTVIPRSFRSKANKREAWAAFYVYKLHQLTYLLDFSVDGYIRRAIDCGHDDLS QLHCSYTSFEVDTGVYSVSSYEASATGTFIEQPNATECDFSPMLTGVAPQVYNFKRLVFSNCNYNLTKLL SLFAVDEFSCNGISPDSIARGCYSTLTVDYFAYPLSMKSYIRPGSAGNIPLYNYKQSFANPTCRVMASVL ANVTITKPHAYGYISKCSRLTGANQDVETPLYINPGEYSICRDFSPGGFSEDGQVFKRTLTQFEGGGLLI GVGTRVPMTDNLQMSFIISVQYGTGTDSVCPMLDLGDSLTITNRLGKCVDYSLYGVTGRGVFQNCTAVGV KQQRFVYDSFDNLVGYYSDDGNYYCVRPCVSVPVSVIYDKSTNLHATLFGSVACEHVTTMMSQFSRLTQS NLRRRDSNIPLQTAVGCVIGLSNNSLVVSDCKLPLGQSLCAVPPVSTFRSYSASQFQLAVLNYTSPIVVT PINSSGFTAAIPTNFSFSVTQEYIETSIQKVTVDCKQYVCNGFTRCEKLLVEYGQFCSKINQALHGANLR QDESVYSLYSNIKTTSTQTLEYGLNGDFNLTLLQVPQIGGSSSSYRSAIEDLLFDKVTIADPGYMQGYDD CMKQGPQSARDLICAQYVSGYKVLPPLYDPNMEAAYTSSLLGSIAGAGWTAGLSSFAAIPFAQSMFYRLN GVGITQQVLSENQKLIANKFNQALGAMQTGFTTSNLAFSKVQDAVNANAQALSKLASELSNTFGAISSSI SDILARLDTVEQDAQIDRLINGRLISLNAFVSQQLVRSETAARSAQLASDKVNECVKSQSKRNGFCGSGT HIVSFVVNAPNGFYFFHVGYVPTNYTNVTAAYGLCNNNNPPLCIAPIDGYFITNQTTTYSVDTEWYYTGS SFYKPEPITQANSRYVSSDVKFDKLENNLPPPLLENSTDVDFKDELEEFFKNVTSHGPNFAEISKINTTL LDLSDEMAMLQEVVKQLNDSYIDLKELGNYTYYNKWPWYVWLGFIAGLVALLLCVFFLLCCTGCGTSCLG KMKCKNCCDSYEEYDVEKIHVH
>QDF43820.1 spike glycoprotein [Coronavirus BtRs-BetaCoV/YN2018A] MKILIFAFLVTLVEAQEGCGIISRKPQPKMAQVSSSRRGVYYNDDIFRSDVLHLTQDYFLPFDSNLTQYF SLNVDSDRYTYFDNPILDFGDGVYFAATEKSNVIRGWIFGSTFDNTTQSAVIVNNSTHIIIRVCNFNLCK EPMYTVSRGTQQSSWVYQSAFNCTYDRVERSFQLDTAPKTGNFKDLREYVFKNRDGFLSVYQTYTAVNLP RGLPIGFSVLRPILKLPFGINITSYRVVMAMFSQTTSNFLPESAAYYVGNLKYTTFMLRFNENGTITDAI DCAQNPLAELKCTIKNFNVSKGIYQTSNFRVSPTQEVVRFPNITNRCPFDKVFNASRFPNVYAWERTKIS DCVADYTVLYNSTSFSTFKCYGVSPSKLIDLCFTSVYADTFLIRSSEVRQVAPGETGVIADYNYKLPDDF TGCVIAWNTAKQDTGHYYYRSHRKTKLKPFERDLSSDDGNGVYTLSTYDFNPNVPVAYQATRVVVLSFEL LNAPATVCGPKLSTQLVKNQCVNFNFNGLKGTGVLTDSSKRFQSFQQFGRDTSDFTDSVRDPQTLEILDI TPCSFGGVSVITPGTNASSEVAVLYQDVNCTDVPTAIRADQLTPAWRVYSTGVNVFQTQAGCLIGAEHVN ASYECDIPIGAGICASYHTASTLRSVGQKSIVAYTMSLGAENSIAYANNSIAIPTNFSISVTTEVMPVSM AKTSVDCTMYICGDSQECSNLLLQYGSFCTQLNRALTGVALEQDKNTQEVFAQVKQMYKTPAIKDFGGFN FSQILPDPSKPTKRSFIEDLLFNKVTLADAGFMKQYGECLGDINARDLICAQKFNGLTVLPPLLTDDMIA AYTAALVSGTATAGWTFGAGAALQIPFAMQMAYRFNGIGVTQNVLYENQKQIANQFNKAISQIQESLTTT STALGKLQDVVNQNAQALNTLVKQLSSNFGAISSVLNDILSRLDKVEAEVQIDRLITGRLQSLQTYVTQQ LIRAAEIRASANLAATKMSECVLGQSKRVDFCGRGYHLMSFPQAAPHGVVFLHVTYVPSQEKNFTTAPAI CHEGKAYFPREGVFVSNGTFWFITQRNFYSPQIITTDNTFVAGNCDVVIGIINNTVYDPLQPELDSFKEE LDKYFKNHTSPDVDLGDISGINASVVNIQKEIDRLNEVAKNLNESLIDLQELGKYEQYIKWPWYVWLGFI AGLIAIVMATILLCCMTSCCSCLKGACSCGSCCKFDEDDSEPVLKGVKLHYT
>AAZ67052.1 spike protein [Bat SARS CoV Rp3/2004] MKILILAFLASLAKAQEGCGIISRKPQPKMAQVSSSRRGVYYNDDIFRSNVLHLTQDYFLPFDSNLTQYF SLNVDSDRFTYFDNPILDFGDGVYFAATEKSNVIRGWIFGSTFDNTTQSAVIVNNSTHIIIRVCNFNLCK EPMYTVSRGAQQSSWVYQSAFNCTYDRVEKSFQLDTAPKTGNFKDLREYVFKNRDGFLSVYQTYTAVNLP RGLPIGFSVLRPILKLPFGINITSYRVVMAMFSQTTSNFLPESAAYYVGNLKYTTFMLSFNENGTITNAI DCAQNPLAELKCTIKNFNVSKGIYQTSNFRVSPTQEVIRFPNITNRCPFDKVFNATRFPNVYAWERTKIS DCVADYTVLYNSTSFSTFKCYGVSPSKLIDLCFTSVYADTFLIRSSEVRQVAPGETGVIADYNYKLPDDF TGCVIAWNTAKQDQGQYYYRSHRKTKLKPFERDLSSDENGVRTLSTYDFYPSVPVAYQATRVVVLSFELL NAPATVCGPKLSTQLVKNQCVNFNFNGLKGTGVLTESSKRFQSFQQFGRDTSDFTDSVRDPQTLEILDIS PCSFGGVSVITPGTNASSEVAVLYQDVNCTDVPAAIHADQLTPAWRVYSTGTNVFQTQAGCLIGAEHVNA SYECDIPIGAGICASYHTASTLRSVGQKSIVAYTMSLGAENSIAYANNSIAIPTNFSISVTTEVMPVSMA KTSVDCTMYICGDSLECSNLLLQYGSFCTQLNRALSGIAIEQDKNTQEVFAQVKQMYKTPAIKDFGGFNF SQILPDPSKPTKRSFIEDLLFNKVTLADAGFMKQYGECLGDISARDLICAQKFNGLTVLPPLLTDEMIAA YTAALVSGTATAGWTFGAGSALQIPFAMQMAYRFNGIGVTQNVLYENQKQIANQFNKAISQIQESLTTTS TALGKLQDVVNQNAQALNTLVKQLSSNFGAISSVLNDILSRLDKVEAEVQIDRLITGRLQSLQTYVTQQL IRAAEIRASANLAATKMSECVLGQSKRVDFCGKGYHLMSFPQAAPHGVVFLHVTYVPSQERNFTTAPAIC HEGKAYFPREGVFVSNGTSWFITQRNFYSPQIITTDNTFVAGSCDVVIGIINNTVYDPLQPELDSFKEEL DKYFKNHTSPDVDLGDISGINASVVNIQKEIDRLNEVAKNLNESLIDLQELGKYEQYIKWPWYVWLGFIA GLIAIVMVTILLCCMTSCCSCLKGACSCGSCCKFDEDDSEPVLKGVKLHYT
>QDF43835.1 spike glycoprotein [Coronavirus BtRs-BetaCoV/YN2018D] MKVLIVLLCLGLVTAQDGCGHISTKPQPLLDKFSSSRRGVYYNDDIFRSDVLHLTQDYFLPFDTNLTRYL SFNMDSATKVYFDNPTLPFGDGIYFAATEKSNVVRGWIFGSTMDNTTQSAIIVNNSTHIIIRVCYFNLCK EPMYAISNEQHYKSWVYQNAYNCTYDRVEQSFQLDTAPQTGNFKDLREYVFKNKDGFLSVYNAYSPIDIP RGLPVGFSVLKPILKLPIGINITSFKVVMSMFSRTTSNFLPEVAAYFVGNLKYSTFMLNFNENGTITDAI DCAQNPLSELKCTIKNFNVSKGIYQTSNFRVSPTHEVIRFPNITNRCPFDKVFNASRFPNVYAWERTKIS DCVADYTVLYNSTSFSTFKCYGVSPSKLIDLCFTSVYADTFLIRSSEVRQVAPGETGVIADYNYKLPDDF TGCVIAWNTAKQDQGQYYYRSSRKTKLKPFERDLTSDENGVRTLSTYDFYPNVPIEYQATRVVVLSFELL NAPATVCGPKLSTGLVKNQCVNFNFNGLRGTGVLTDSSKRFQSFQQFGRDTSDFTDSVRDPQTLEILDIT PCSFGGVSVITPGTNASSEVAVLYQDVNCTDVPTAIRADQLTPAWRVYSTGINVFQTQAGCLIGAEHVNA SYECDIPIGAGICASYHTASTLRSVGQKSIVAYTMSLGAENSIAYANNSIAIPTNFSISVTTEVMPVSMS KTSVDCTMYICGDSQECSNLLLQYGSFCTQLNRALTGIAIEQDKNTQEVFAQVKQMYKTPAIKDFGGFNF SQILPDPSKPTKRSFIEDLLFNKVTLADAGFMKQYGECLGDINARDLICAQKFNGLTVLPPLLTDDMIAA YTAALVSGTATAGWTFGAGAALQIPFAMQMAYRFNGIGVTQNVLYENQKQIANQFNKAISQIQESLTTTS TALGKLQDVVNQNAQALNTLVKQLSSNFGAISSVLNDILSRLDKVEAEVQIDRLITGRLQSLQTYVTQQL IRAAEIRASANLAATKMSECVLGQSKRVDFCGRGYHLMSFPQAAPHGVVFLHVTYVPSQEKNFTTAPAIC HEGKAYFPREGVFVSNGTSWFITQRNFYSPQIITTDNTFVAGSCDVVIGIINNTVYDPLQPELDSFKEEL DKYFKNHTSPDVDLGDISGINASVVNIQKEIDRLNEVAKNLNESLIDLQELGKYEQYIKWPWYVWLGFIA GLIAIVMATILLCCMTSCCSCLKGACSCGSCCKFDEDDSEPVLKGVKLHYT
>ALK02457.1 spike protein [SARS-like coronavirus WIV16] MFIFLFFLTLTSGSDLESCTTFDDVQAPNYPQHSSSRRGVYYPDEIFRSDTLYLTQDLFLPFYSNVTGFH TINHRFDNPVIPFKDGVYFAATEKSNVVRGWVFGSTMNNKSQSVIIINNSTNVVIRACNFELCDNPFFAV SKPTGTQTHTMIFDNAFNCTFEYISDSFSLDVAEKSGNFKHLREFVFKNKDGFLYVYKGYQPIDVVRDLP SGFNILKPIFKLPLGINITNFRAILTAFLPAQDTWGTSAAAYFVGYLKPATFMLKYDENGTITDAVDCSQ NPLAELKCSVKSFEIDKGIYQTSNFRVAPSKEVVRFPNITNLCPFGEVFNATTFPSVYAWERKRISNCVA DYSVLYNSTSFSTFKCYGVSATKLNDLCFSNVYADSFVVKGDDVRQIAPGQTGVIADYNYKLPDDFTGCV LAWNTRNIDATQTGNYNYKYRSLRHGKLRPFERDISNVPFSPDGKPCTPPAFNCYWPLNDYGFYITNGIG YQPYRVVVLSFELLNAPATVCGPKLSTDLIKNQCVNFNFNGLTGTGVLTPSSKRFQPFQQFGRDVLDFTD SVRDPKTSEILDISPCSFGGVSVITPGTNTSSEVAVLYQDVNCTDVPVAIHADQLTPSWRVYSTGNNVFQ TQAGCLIGAEHVDTSYECDIPIGAGICASYHTVSSLRSTSQKSIVAYTMSLGADSSIAYSNNTIAIPTNF SISITTEVMPVSMAKTSVDCNMYICGDSTECANLLLQYGSFCTQLNRALSGIAVEQDRNTREVFAQVKQM YKTPTLKDFGGFNFSQILPDPLKPTKRSFIEDLLFNKVTLADAGFMKQYGECLGDINARDLICAQKFNGL TVLPPLLTDDMIAAYTAALVSGTATAGWTFGAGAALQIPFAMQMAYRFNGIGVTQNVLYENQKQIANQFN KAISQIQESLTTTSTALGKLQDVVNQNAQALNTLVKQLSSNFGAISSVLNDILSRLDKVEAEVQIDRLIT GRLQSLQTYVTQQLIRAAEIRASANLAATKMSECVLGQSKRVDFCGKGYHLMSFPQAAPHGVVFLHVTYV PSQERNFTTAPAICHEGKAYFPREGVFVFNGTSWFITQRNFFSPQIITTDNTFVSGSCDVVIGIINNTVY DPLQPELDSFKEELDKYFKNHTSPDVDLGDISGINASVVNIQKEIDRLNEVAKNLNESLIDLQELGKYEQ YIKWPWYVWLGFIAGLIAIVMVTILLCCMTSCCSCLKGACSCGSCCKFDEDDSEPVLKGVKLHYT
>ABD75323.1 spike protein [Bat SARS CoV Rf1/2004] MKILIFAFLVTLVKAQEGCGVINLRTQPKLTQVSSSRRGVYYNDDIFRSDVLHLTQDYFLPFHSNLTQYF SLNIESDKIVYFDNPILKFGDGVYFAATEKSNVIRGWVFGSTFDNTTQSAIIVNNSTHIIIRVCYFNLCK DPMYTVSAGTQKSSWVYQSAFNCTYDRVEKSFQLDTSPKTGNFTDLREFVFKNRDGFFTAYQTYTPVNLL RGLPSGLSVLKPILKLPFGINITSFRVVMAMFSKTTSNYVPESAAYYVGNLKQSTFMLSFNQNGTIVDAV DCSQDPLAELKCTTKSFNVSKGIYQTSNFRVSPVTEVVRFPNITNLCPFDKVFNATRFPSVYAWERTKIS DCVADYTVFYNSTSFSTFNCYGVSPSKLIDLCFTSVYADTFLIRFSEVRQVAPGQTGVIADYNYKLPDDF TGCVIAWNTAKQDVGSYFYRSHRSSKLKPFERDLSSEENGVRTLSTYDFNQNVPLEYQATRVVVLSFELL NAPATVCGPKLSTSLVKNQCVNFNFNGFKGTGVLTDSSKTFQSFQQFGRDASDFTDSVRDPQTLRILDIS PCSFGGVSVITPGTNTSSAVAVLYQDVNCTDVPRTIQADQLAPSWRVYTTGPYVFQTQAGCLIGAEHVNA SYQCDIPIGAGICASYHTASHLRSTGQKSIVAYTMSLGAENSVAYANNSIAIPTNFSISVTTEVMPVSMA KTSVDCTMYICGDSLECSNLLLQYGSFCTQLNRALSGIAVEQDKNTQEVFAQVKQMYKTPTIRDFGGFNF SQILPDPLKPTKRSFIEDLLYNKVTLADAGFMKQYADCLGGINARDLICAQKFNGLTVLPPLLTDDMIAA YTAALISGTATAGWTFGAGAALQIPFAMQMAYRFNGIGVTQNVLYENQKQIANQFNKAITQIQESLTTTS TALGKLQDVVNQNAQALNTLVKQLSSNFGAISSALNDILSRLDKVEAEVQIDRLITGRLQSLQTYVTQQL IRAAEIRASANLAATKMSECVLGQSKRVDFCGKGYHLMSFPQSAPHGVVFLHVTYVPSQEKNFTTAPAIC HEGKAYFPREGVFVSNGSSWFITQRNFYSPQIITTDNTFVAGSCDVVIGIINNTVYDPLQPELDSFKQEL DKYFKNHTSPDVDLGDISGINASVVDIQKEIDRLNEVAKNLNESLIDLQELGKYEQYIKWPWYVWLGFIA GLVGLFMAIILLCYFTSCCSCCKGMCSCGSCCRFDEDDSEPVLKGVKLHYT
>AAS10463.1 spike glycoprotein [SARS coronavirus GD03T0013] MFIFLLFLTLTSGSDLDRCTTFDDVQAPNYTQHTSSMRGVYYPDEIFRSDTLYLTQDLFLPFYSNVTGFH TINHTFDDPVIPFKDGIYFAATEKSNVVRGWVFGSTMNNKSQSVIIINNSTNVVIRACNFELCDNPFFVV SKPMGTRTHTMIFDNAFNCTFEYISDAFSLDVSEKSGNFKHLREFVFKNKDGFLYVYKGYQPIDVVRDLP SGFNTLKPIFKLPLGINITNFRAILTAFSPAQDTWGTSAAAYFVGYLKPTTFMLKYDENGTITDAVDCSQ NPLAELKCSVKSFEIDKGIYQTSNFRVVPSGDVVRFPNITNLCPFGEVFNATKFPSVYAWERKRISNCVA DYSVLYNSTSFSTFKCYGVSATKLNDLCFSNVYADSFVVKGDDVRQIAPGQTGVIADYNYKLPDDFMGCV LAWNTRNIDATSTGNYNYKYRYLRHGKLRPFERDISNVPFSPDGKPCTPPAPNCYWPLNGYGFYTTSGIG YQPYRVVVLSFELLNAPATVCGPKLSTDLIKNQCVNFNFNGLTGTGVLTPSSKRFQPFQQFGRDVSDFTD SVRDPKTSEILDISPCSFGGVSVITPGTNASSEVAVLYQDVNCTDVSTLIHAEQLTPAWRIYSTGNNVFQ TQAGCLIGAEHVDTSYECDIPIGAGICASYHTVSSLRSTSQKSIVAYTMSLGADSSIAYSNNTIAIPTNF SISITTEVMPVSMAKTSVDCNMYICGDSTECANLLLQYGSFCRQLNRALSGIAAEQDRNTREVFVQVKQM YKTPTLKDFGGFNFSQILPDPLKPTKRSFIEDLLFNKVTLADAGFMKQYGECLGDINARDLICAQKFNGL TVLPPLLTDDMIAAYTAALVSGTATAGWTFGAGAALQIPFAMQMAYRFNGIGVTQNVLYENQKQIANQFN KAISQIQESLTTTSTALGKLQDVVNQNAQALNTLVKQLSSNFGAISSVLNDILSRLDKVEAEVQIDRLIT GRLQSLQTYVTQQLIRAAEIRASANLAATKMSECVLGQSKRVDFCGKGYHLMSFPQAAPHGVVFLHVTYV PSQERNFTTAPAICHEGKAYFPREGVFVFNGTSWFITQRNFFSPQIITTDNTFVSGNCDVVIGIINNTVY DPLQPELDSFKEELDKYFKNHTSPDVDLGDISGINASVVNIQEEIDRLNEVAKNLNESLIDLQELGKYEQ YIKWPWYVWLGFIAGLIAIVMVTILLCCMTSCCSCLKGACSCGSCCKFDEDDSEPVLKGVKLHYT
>AAP13567.1 putative E2 glycoprotein precursor [SARS coronavirus CUHK-W1] MFIFLLFLTLTSGSDLDRCTTFDDVQAPNYTQHTSSMRGVYYPDEIFRSDTLYLTQDLFLPFYSNVTGFH TINHTFDNPVIPFKDGIYFAATEKSNVVRGWVFGSTMNNKSQSVIIINNSTNVVIRACNFELCDNPFFAV SKPMGTQTHTMIFDNAFNCTFEYISDAFSLDVSEKSGNFKHLREFVFKNKDGFLYVYKGYQPIDVVRDLP SGFNTLKPIFKLPLGINITNFRAILTAFSPAQDTWGTSAAAYFVGYLKPTTFMLKYDENGTITDAVDCSQ NPLAELKCSVKSFEIDKGIYQTSNFRVVPSGDVVRFPNITNLCPFGEVFNATKFPSVYAWERKKISNCVA DYSVLYNSTFFSTFKCYGVSATKLNDLCFSNVYADSFVVKGDDVRQIAPGQTGVIADYNYKLPDDFMGCV LAWNTRNIDATSTGNYNYKYRYLRHGKLRPFERDISNVPFSPDGKPCTPPALNCYWPLNDYGFYTTTGIG YQPYRVVVLSFELLNAPATVCGPKLSTDLIKNQCVNFNFNGLTGTGVLTPSSKRFQPFQQFGRDVSDFTD SVRDPKTSEILDISPCSFGGVSVITPGTNASSEVAVLYQDVNCTDVSTAIHADQLTPAWRIYSTGNNVFQ TQAGCLIGAEHVDTSYECDIPIGAGICASYHTVSLLRSTSQKSIVAYTMSLGADSSIAYSNNTIAIPTNF SISITTEVMPVSMAKTSVDCNMYICGDSTECANLLLQYGSFCTQLNRALSGIAAEQDRNTREVFAQVKQM YKTPTLKYFGGFNFSQILPDPLKPTKRSFIEDLLFNKVTLADAGFMKQYGECLGDINARDLICAQKFNGL TVLPPLLTDDMIAAYTAALVSGTATAGWTFGAGAALQIPFAMQMAYRFNGIGVTQNVLYENQKQIANQFN KAISQIQESLTTTSTALGKLQDVVNQNAQALNTLVKQLSSNFGAISSVLNDILSRLDKVEAEVQIDRLIT GRLQSLQTYVTQQLIRAAEIRASANLAATKMSECVLGQSKRVDFCGKGYHLMSFPQAAPHGVVFLHVTYV PSQERNFTTAPAICHEGKAYFPREGVFVFNGTSWFITQRNFFSPQIITTDNTFVSGNCDVVIGIINNTVY DPLQPELDSFKEELDKYFKNHTSPDVDLGDISGINASVVNIQKEIDRLNEVAKNLNESLIDLQELGKYEQ YIKWPWYVWLGFIAGLIAIVMVTILLCCMTSCCSCLKGACSCGSCCKFDEDDSEPVLKGVKLHYT
>AVP78031.1 spike protein [Bat SARS-like coronavirus] MLFFLFLQFALVNSQCVNLTGRTPLNPNYTNSSQRGVYYPDTIYRSDTLVLSQGYFLPFYSNVSWYYSLT TNNAATKRTDNPILDFKDGIYFAATEHSNIIRGWIFGTTLDNTSQSLLIVNNATNVIIKVCNFDFCYDPY LSGYYHNNKTWSIREFAVYSSYANCTFEYVSKSFMLNISGNGGLFNTLREFVFRNVDGHFKIYSKFTPVN LNRGLPTGLSVLQPLVELPVSINITKFRTLLTIHRGDPMPNNGWTAFSAAYFVGYLKPRTFMLKYNENGT ITDAVDCALDPLSETKCTLKSLTVQKGIYQTSNFRVQPTQSVVRFPNITNVCPFHKVFNATRFPSVYAWE RTKISDCIADYTVFYNSTSFSTFKCYGVSPSKLIDLCFTSVYADTFLIRFSEVRQVAPGQTGVIADYNYK LPDDFTGCVIAWNTAKQDVGNYFYRSHRSTKLKPFERDLSSDENGVRTLSTYDFNPNVPLEYQATRVVVL SFELLNAPATVCGPKLSTQLVKNQCVNFNFNGLKGTGVLTDSSKRFQSFQQFGKDASDFIDSVRDPQTLE ILDITPCSFGGVSVITPGTNTSLEVAVLYQDVNCTDVPTTIHADQLTPAWRIYATGTNVFQTQAGCLIGA EHVNASYECDIPIGAGICASYHTASILRSTSQKAIVAYTMSLGAENSIAYANNSIAIPTNFSISVTTEVM PVSMAKTSVDCTMYICGDSIECSNLLLQYGSFCTQLNRALSGIAIEQDKNTQEVFAQVKQIYKTPPIKDF GGFNFSQILPDPSKPSKRSFIEDLLFNKVTLADAGFIKQYGDCLGGISARDLICAQKFNGLTVLPPLLTD EMIAAYTAALISGTATAGWTFGAGAALQIPFAMQMAYRFNGIGVTQNVLYENQKLIANQFNSAIGKIQES LTSTASALGKLQDVVNQNAQALNTLVKQLSSNFGAISSVLNDILSRLDKVEAEVQIDRLITGRLQSLQTY VTQQLIRAAEIRASANLAATKMSECVLGQSKRVDFCGKGYHLMSFPQSAPHGVVFLHVTYIPSQEKNFTT APAICHEGKAHFPREGVFVSNGTHWFVTQRNFYEPKIITTDNTFVSGNCDVVIGIINNTVYDPLQPELDS FKEELDKYFKNHTSPDIDLGDISGINASVVNIQKEIDRLNEVARNLNESLIDLQELGKYEQYIKWPWYVW LGFIAGLIAIVMVTILLCCMTSCCSCLKGCCSCGSCCKFDEDDSEPVLKGVKLHYT
>ABD75332.1 spike protein [Bat SARS CoV Rm1/2004] MKVLIFALLFSLAKAQEGCGIISRKPQPKMEKVSSSRRGVYYNDDIFRSDVLHLTQDYFLPFDSNLTQYF SLNIDSNKYTYFDNPILDFGDGVYFAATEKSNVIRGWIFGSSFDNTTQSAIIVNNSTHIIIRVCNFNLCK EPMYTVSKGTQQSSWVYQSAFNCTYDRVEKSFQLDTAPKTGNFKDLREYVFKNKGGFLRVYQTYTAVNLP RGFPAGFSVLRPILKLPFGINITSYRVVMTMFSQFNSNFLPESAAYYVGNLKYTTFMLSFNENGTITDAV DCSQNPLAELKCTIKNFNVSKGIYQTSNFRVTPTQEVVRFPNITNRCPFDKVFNASRFPNVYAWERTKIS DCVADYTVLYNSTSFSTFKCYGVSPSKLIDLCFTSVYADTFLIRSSEVRQVAPGETGVIADYNYKLPDDF TGCVIAWNTAQQDQGQYYYRSYRKEKLKPFERDLSSDENGVYTLSTYDFYPSIPVEYQATRVVVLSFELL NAPATVCGPKLSTQLVKNQCVNFNFNGLRGTGVLTTSSKRFQSFQQFGRDTSDFTDSVRDPQTLEILDIS PCSFGGVSVITPGTNASSEVAVLYQDVNCTDVPTSIHADQLTPAWRVYSTGVNVFQTQAGCLIGAEHVNA SYECDIPIGAGICASYHTASVLRSTGQKSIVAYTMSLGAENSIAYANNSIAIPTNFSISVTTEVMPVSIA KTSVDCTMYICGDSLECSNLLLQYGSFCTQLNRALTGIAIEQDKNTQEVFAQVKQMYKTPAIKDFGGFNF SQILPDPSKPTKRSFIEDLLFNKVTLADAGFMKQYGECLGDISARDLICAQKFNGLTVLPPLLTDEMIAA YTAALVSGTATAGWTFGAGSALQIPFAMQMAYRFNGIGVTQNVLYENQKQIANQFNKAISQIQESLTTTS TALGKLQDVVNQNAQALNTLVKQLSSNFGAISSVLNDILSRLDKVEAEVQIDRLITGRLQSLQTYVTQQL IRAAEIRASANLAATKMSECVLGQSKRVDFCGKGYHLMSFPQAAPHGVVFLHVTYVPSQERNFTTAPAIC HEGKAYFPREGVFVSNGTSWFITQRNFYSPQIITTDNTFVAGNCDVVIGIINNTVYDPLQPELDSFKEEL DKYFKNHTSPDVDLGDISGINASVVNIQKEIDRLNEVAKNLNESLIDLQELGKYEQYIKWPWYVWLGFIA GLIAIVMVTILLCCMTSCCSCLKGACSCGSCCKFDEDDSEPVLKGVKLHYT
>AGZ48818.1 spike protein [Bat SARS-like coronavirus Rs3367] MKLLVLVFATLVSSYTIEKCLDFDDRTPPANTQFLSSHRGVYYPDDIFRSNVLHLVQDHFLPFDSNVTRF ITFGLNFDNPIIPFKDGIYFAATEKSNVIRGWVFGSTMNNKSQSVIIMNNSTNLVIRACNFELCDNPFFV VLKSNNTQIPSYIFNNAFNCTFEYVSKDFNLDLGEKPGNFKDLREFVFRNKDGFLHVYSGYQPISAASGL PTGFNALKPIFKLPLGINITNFRTLLTAFPPRPDYWGTSAAAYFVGYLKPTTFMLKYDENGTITDAVDCS QNPLAELKCSVKSFEIDKGIYQTSNFRVAPSKEVVRFPNITNLCPFGEVFNATTFPSVYAWERKRISNCV ADYSVLYNSTSFSTFKCYGVSATKLNDLCFSNVYADSFVVKGDDVRQIAPGQTGVIADYNYKLPDDFTGC VLAWNTRNIDATQTGNYNYKYRSLRHGKLRPFERDISNVPFSPDGKPCTPPAFNCYWPLNDYGFYITNGI GYQPYRVVVLSFELLNAPATVCGPKLSTDLIKNQCVNFNFNGLTGTGVLTPSSKRFQPFQQFGRDVSDFT DSVRDPKTSEILDISPCSFGGVSVITPGTNTSSEVAVLYQDVNCTDVPVAIHADQLTPSWRVYSTGNNVF QTQAGCLIGAEHVDTSYECDIPIGAGICASYHTVSSLRSTSQKSIVAYTMSLGADSSIAYSNNTIAIPTN FSISITTEVMPVSMAKTSVDCNMYICGDSTECANLLLQYGSFCTQLNRALSGIAVEQDRNTREVFAQVKQ MYKTPTLKDFGGFNFSQILPDPLKPTKRSFIEDLLFNKVTLADAGFMKQYGECLGDINARDLICAQKFNG LTVLPPLLTDDMIAAYTAALVSGTATAGWTFGAGAALQIPFAMQMAYRFNGIGVTQNVLYENQKQIANQF NKAISQIQESLTTTSTALGKLQDVVNQNAQALNTLVKQLSSNFGAISSVLNDILSRLDKVEAEVQIDRLI TGRLQSLQTYVTQQLIRAAEIRASANLAATKMSECVLGQSKRVDFCGKGYHLMSFPQAAPHGVVFLHVTY VPSQERNFTTAPAICHEGKAYFPREGVFVFNGTSWFITQRNFFSPQIITTDNTFVSGSCDVVIGIINNTV YDPLQPELDSFKEELDKYFKNHTSPDVDLGDISGINASVVNIQKEINRLNEVAKNLNESLIDLQELGKYE QYIKWPWYVWLGFIAGLIAIVMVTILLCCMTSCCSCLKGACSCGSCCKFDEDDSEPVLKGVKLHYT
>AGZ48806.1 spike protein [Bat SARS-like coronavirus RsSHC014] MKLLVLVFATLVSSYTIEKCLDFDDRTPPANTQFLSSHRGVYYPDDIFRSNVLHLVQDHFLPFDSNVTRF ITFGLNFDNPIIPFRDGIYFAATEKSNVIRGWVFGSTMNNKSQSVIIMNNSTNLVIRACNFELCDNPFFV VLKSNNTQIPSYIFNNAFNCTFEYVSKDFNLDLGEKPGNFKDLREFVFRNKDGFLHVYSGYQPISAASGL PTGFNALKPIFKLPLGINITNFRTLLTAFPPRPDYWGTSAAAYFVGYLKPTTFMLKYDENGTITDAVDCS QNPLAELKCSVKSFEIDKGIYQTSNFRVAPSKEVVRFPNITNLCPFGEVFNATTFPSVYAWERKRISNCV ADYSVLYNSTSFSTFKCYGVSATKLNDLCFSNVYADSFVVKGDDVRQIAPGQTGVIADYNYKLPDDFLGC VLAWNTNSKDSSTSGNYNYLYRWVRRSKLNPYERDLSNDIYSPGGQSCSAVGPNCYNPLRPYGFFTTAGV GHQPYRVVVLSFELLNAPATVCGPKLSTDLIKNQCVNFNFNGLTGTGVLTPSSKRFQPFQQFGRDVSDFT DSVRDPKTSEILDISPCSFGGVSVITPGTNTSSEVAVLYQDVNCTDVPVAIHADQLTPSWRVYSTGNNVF QTQAGCLIGAEHVDTSYECDIPIGAGICASYHTVSSLRSTSQKSIVAYTMSLGADSSIAYSNNTIAIPTN FSISITTEVMPVSMAKTSVDCNMYICGDSTECANLLLQYGSFCTQLNRALSGIAVEQDRNTREVFAQVKQ MYKTPTLKDFGGFNFSQILPDPLKPTKRSFIEDLLFNKVTLADAGFMKQYGECLGDINARDLICAQKFNG LTVLPPLLTDDMIAAYTAALVSGTATAGWTFGAGAALQIPFAMQMAYRFNGIGVTQNVLYENQKQIANQF NKAISQIQESLTTTSTALGKLQDVVNQNAQALNTLVKQLSSNFGAISSVLNDILSRLDKVEAEVQIDRLI TGRLQSLQTYVTQQLIRAAEIRASANLAATKMSECVLGQSKRVDFCGKGYHLMSFPQAAPHGVVFLHVTY VPSQERNFTTAPAICHEGKAYFPREGVFVFNGTSWFITQRNFFSPQIITTDNTFVSGSCDVVIGIINNTV YDPLQPELDSFKEELDKYFKNHTSPDVDLGDISGINASVVNIQKEIDRLNEVAKNLNESLIDLQELGKYE QYIKWPWYVWLGFIAGLIAIVMVTILLCCMTSCCSCLKGACSCGSCCKFDEDDSEPVLKGVKLHYT
>AGZ48806.1 spike protein [Bat SARS-like coronavirus RsSHC014] MKLLVLVFATLVSSYTIEKCLDFDDRTPPANTQFLSSHRGVYYPDDIFRSNVLHLVQDHFLPFDSNVTRF ITFGLNFDNPIIPFRDGIYFAATEKSNVIRGWVFGSTMNNKSQSVIIMNNSTNLVIRACNFELCDNPFFV VLKSNNTQIPSYIFNNAFNCTFEYVSKDFNLDLGEKPGNFKDLREFVFRNKDGFLHVYSGYQPISAASGL PTGFNALKPIFKLPLGINITNFRTLLTAFPPRPDYWGTSAAAYFVGYLKPTTFMLKYDENGTITDAVDCS QNPLAELKCSVKSFEIDKGIYQTSNFRVAPSKEVVRFPNITNLCPFGEVFNATTFPSVYAWERKRISNCV ADYSVLYNSTSFSTFKCYGVSATKLNDLCFSNVYADSFVVKGDDVRQIAPGQTGVIADYNYKLPDDFLGC VLAWNTNSKDSSTSGNYNYLYRWVRRSKLNPYERDLSNDIYSPGGQSCSAVGPNCYNPLRPYGFFTTAGV GHQPYRVVVLSFELLNAPATVCGPKLSTDLIKNQCVNFNFNGLTGTGVLTPSSKRFQPFQQFGRDVSDFT DSVRDPKTSEILDISPCSFGGVSVITPGTNTSSEVAVLYQDVNCTDVPVAIHADQLTPSWRVYSTGNNVF QTQAGCLIGAEHVDTSYECDIPIGAGICASYHTVSSLRSTSQKSIVAYTMSLGADSSIAYSNNTIAIPTN FSISITTEVMPVSMAKTSVDCNMYICGDSTECANLLLQYGSFCTQLNRALSGIAVEQDRNTREVFAQVKQ MYKTPTLKDFGGFNFSQILPDPLKPTKRSFIEDLLFNKVTLADAGFMKQYGECLGDINARDLICAQKFNG LTVLPPLLTDDMIAAYTAALVSGTATAGWTFGAGAALQIPFAMQMAYRFNGIGVTQNVLYENQKQIANQF NKAISQIQESLTTTSTALGKLQDVVNQNAQALNTLVKQLSSNFGAISSVLNDILSRLDKVEAEVQIDRLI TGRLQSLQTYVTQQLIRAAEIRASANLAATKMSECVLGQSKRVDFCGKGYHLMSFPQAAPHGVVFLHVTY VPSQERNFTTAPAICHEGKAYFPREGVFVFNGTSWFITQRNFFSPQIITTDNTFVSGSCDVVIGIINNTV YDPLQPELDSFKEELDKYFKNHTSPDVDLGDISGINASVVNIQKEIDRLNEVAKNLNESLIDLQELGKYE QYIKWPWYVWLGFIAGLIAIVMVTILLCCMTSCCSCLKGACSCGSCCKFDEDDSEPVLKGVKLHYT
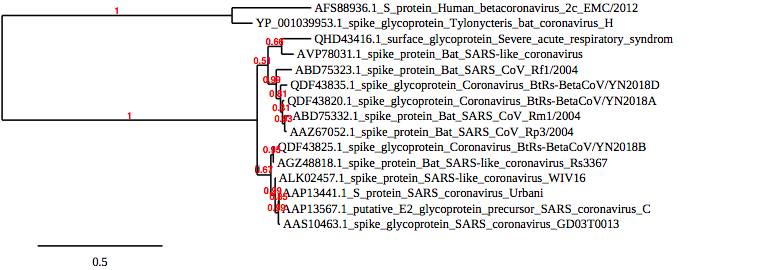
- tab 6. Tree Rendering was then clicked after going back to the previous screen, and the phylogenetic tree of the five sequences was seen.
- comparison of the tree to the multiple sequence alignment. and the differences in the sequences:
The class sequence alignment is very similar to the phylogenetic tree above. For example, both (QDF43825.1) and (AGZ48818.1) are closely related in the phylogeny tree and when looking at their sequences they are very similar too. Further, when we compare the sequence of (QDF43825.1) to (ALK02457.1) we find similarities between the two but the sequences are more different from each other than (QDF43825.1) and (AGZ48818.1) are. This makes sense when looking at the phylogenetic tree as (QDF43825.1) and (ALK02457.1) branch from the same point but are further away from each other than (QDF43825.1)and (AGZ48818.1).
- alignment compared to Figure 3 of Wan et al (2020) paper:
The sequence shown in figure 3 of the paper is conserved, there’s much more prevalence in the class sequence than there’s in the paper. The sequence provided by the article has different amino acids when compared to the class sequence.
- The tree compared to Figure 2 of Wan et al (2020) paper:
The tree from the article used genomic sequences however the tree from the class sequence included spike proteins sequences. Also, both trees have two branches. The phylogenetic tree from the paper seems to have one outgroup including one sequence (BtSCoV PDF2386) compared to the phylogenetic tree obtained from the class sequence had an outgroup including two sequences (AAP13567.1) and (AAS10463.1). The tree from the class sequence included all the sequences but from the paper, it included only part of it.
- It was then interpreted if there was enough information provided by Wan et al (2020) in their paper for their analysis to be reproduced:
the paper did not provide sufficient information for it to be replicated. The methods section was very limited and it's hard to tell how they found the information that they did from just looking at the methods. Also, there's not much information about why they chose to include some results but not the other. Therefore, I don't believe this paper is replicable.
Conclusion
In this exercise, I was able to Learn about obtaining sequence data and comparing it using multiple sequence alignments which is an important skill for a biologist to have in order to be able to scientifically analyze sequences and the phylogenetic tree which tell a lot about the origin of which species, proteins, viruses, etc. come from and how they are related thereby providing essential information that could help me when conducting my own research.
Acknowledgements
- GenBank for the SARS Coronavirus Urbani DNA sequences and information related to assigned
- copied and modified the protocol shown on the Week 4 page
- I acknowledge the instructions from week 4[1]
- I discussed questions with My partner Kam Taghizadeh.
- Phylogeny.fr used to generate phylogenetic trees
- Referred to data in the article et al (2020) paper
- Dr. Kam D. Dahlquist, Ph.D. helped me in understanding the homework.
"Except for what is noted above, this individual journal entry was completed by me and not copied from another source"Falghane (talk) 17:19, 1 October 2020 (PDT)
References
- OpenWetWare. (2020). BIOL368/F20:Week 1. Retrieved October 1, 2020, from https://openwetware.org/wiki/BIOL368/F20:Week_4
- OpenWetWare. (2020). BIOL368/F20:Week 4. Retrieved October 1, 2020, from https://openwetware.org/wiki/BIOL368/F20:Week_1
- http://www.phylogeny.fr/simple_phylogeny.cgi
- Ncbi.nlm.nih.gov. 2020. Genbank Overview. [online] Available at: https://www.ncbi.nlm.nih.gov/genbank/ [Accessed 2 October 2020].
- http://www.phylogeny.fr/
- Wan, Y., Shang, J., Graham, R., Baric, R. and Li, F., 2020. Receptor Recognition By The Novel Coronavirus From Wuhan: An Analysis Based On Decade-Long Structural Studies Of SARS Coronavirus.