User:Mbennie/Notebook/Lab Notebook/Notebook/2007/08/16
From OpenWetWare
- PCR
- Template: 40ul PCR Supermix, .4ul of each primer, and .4ul of PCR purification product
- B: Part 1 of IgA beta-core IgAb-F and Mut_Pst1a-R
- C: Part 2 of IgA beta-core Mut_Pst1a-F and Mut2_Pst1b-R
- D: Part 3 of IgA beta-core Mut2_Pst1b-F and IgAb-R
- Same thermocycler protocol as 8.1.2007
- Template: 40ul PCR Supermix, .4ul of each primer, and .4ul of PCR purification product
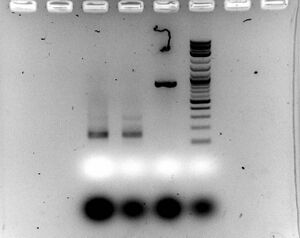
- Gel
- Ran 1.5% gel for 30 minutes at 100V with samples (5ul sample with 2ul of loading dye)
- Interesting: it appears that the PCR is the issue as the 200bp fragment was amplified solely from the correct length DNA fragment
- Digest
- B: 5ul B DNA, 2ul NEB4, .5ul Sap1, rest water (20ul rxn)
- C: 5ul C DNA, 2ul NEB4, .5ul Sap1, rest water (20ul rxn)
- D: 5ul D DNA, 2ul NEB4, .5ul Sap1, rest water (20ul rxn)
- Thermocycler protocol: 2hr@37C, 20mins@80C
- Ligation
- C: 6ul of C digest DNA, 7ul water, 1.5ul ligase buffer, .5ul ligase
- D: 6ul of D digest DNA, 7ul water, 1.5ul ligase buffer, .5ul ligase
- C + D: 6ul of C digest DNA, 6ul of D digest DNA, 1ul water, 1.5ul ligase buffer, .5ul ligase
- B + C + D: 6ul of each digest DNA, 2ul ligase buffer, .5ul ligase
- Protocol: 30mins@roomtemp,10mins@65C
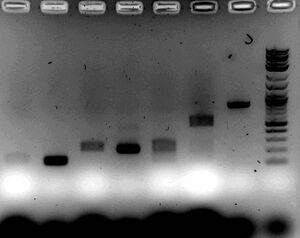
- Gel
- Ran 1.5% gel for 30 minutes at 100V with samples
- Ligations were completely used and mixed with about 5ul of loading dye
- PCR products were used in normal proportions (5ul DNA, 2ul dye)
- Looks like there is a ligation issue with part D
- Ran 1.5% gel for 30 minutes at 100V with samples
- Oligos
- 6His_Tag-F
- 6His_Tag-R
- FLAG_Tag-F
- FLAG_Tag-R
- Myc_Tag-F
- Myc_Tag-R
- HA_Tag-F
- HA_Tag-R
- GCN4-R2
- Resuspended at 50uM
- PCR
- Template: 40ul PCR Supermix, .8ul primers (.4 primers for GCN4), .8ul DNA (only GCN4)
- 6His Tag: 6His_Tag-F and 6His_Tag-R
- FLAG Tag: FLAG_Tag-F and FLAG_Tag-R
- Myc Tag: Myc_Tag-F and Myc_Tag-R
- HA Tag: HA_Tag-F and HA_Tag-R
- GCN4 Leucine Zipper: GCN4-F and GCN4-R2
- Same protocol as 8.1.2007
- Template: 40ul PCR Supermix, .8ul primers (.4 primers for GCN4), .8ul DNA (only GCN4)
- PCR Purification
- Used MinElute columns to PCR purify tubes B, C, and D
- Eluted in 10ul of water
- Digest
- Template: 3ul DNA (each), 2ul NEB2, .5ul Ear1, rest water (20ul rxn)
- B+C
- C+D
- B+C+D
- Thermocycler protocol: 1hr@37C, 20mins@80C
- Template: 3ul DNA (each), 2ul NEB2, .5ul Ear1, rest water (20ul rxn)
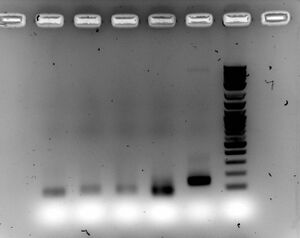
- Gel
- Ran 1.5% gel for 30 minutes at 100V with samples (5ul sample with 2ul of loading dye)
- Everything looks good, will ligate into vector tomorrow
- Ligation
- Performed on digest tube above
- Template (50ul rxns): 20ul digest, 5ul ligase buffer, .5ul ligase, rest water
- Thermocycler protocol: 1hr@16C,10mins@65C
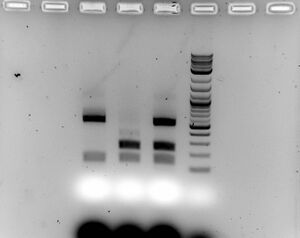
- Gel
- Ran 1.5% gel for 30 minutes at 100V with samples (15ul sample with 5ul of loading dye)
- Looks promising, although very low ligation efficiency
- PCR
- Template: 40ul PCR Supermix, .4ul of each primer, and 1ul of ligation product
- B+C: IgAb-F and Mut2_Pst1b-R
- C+D: Mut_Pst1a-F and IgAb-R
- B+C+D:IgAb-F and IgAb-R
- B+C+D: BB_f and BB_r
- Template: 40ul PCR Supermix, .4ul of each primer, and 1ul of ligation product
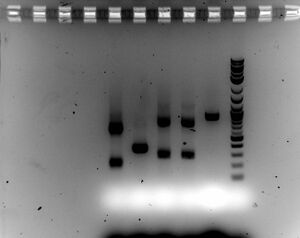
- Gel
- Ran 1.5% gel for 30 minutes at 100V with samples (5ul sample with 2ul of loading dye)
- A closer inspection reveals that cutting and ligating were successful (although the full construct was missing part C)
- The rouge bands can be explained by incorrect hybridization of primers about 100bp from the end of part B
- The two ligations involving two parts look good and are fairly clean -> see if adding last part works