Nicolette S. Harmon Week 9
From OpenWetWare
Jump to navigationJump to search
HIV Structure Project
This is the newer version Media:HIVstructureNickiSam.ppt
Media:HIVstructureNickiSam.pptx
FINAL= Media:HIVstructureNickiSamFINAL.ppt
Methods
- The subjects that we selected for this project are subjects 2,8,12,13.
- We picked subjects 2,12,13 because they are the nonprogressor types and subject 8 is the moderate progressor that we will be using as a comparison to the other three subjects.
- To retrieve the nucleotide and amino acid sequences needed for these subjects, I used Bedrock.
- We randomly selected to use visit 3 as for our nucleotide and amino acid sequences for the nonprogressors since we only need to use one visit due to the fact there shouldn't be large changes in the sequence.
- For subject 8 we used the nucleotide and amino acid sequences from the first and last visit since we are expecting there to be a large change over the course of the Markham study.
- These sequences were then uploaded onto the Biology Workbench site.
- Then, I used the Multiple Sequence Alignment tool selecting all the sequences to compare them.
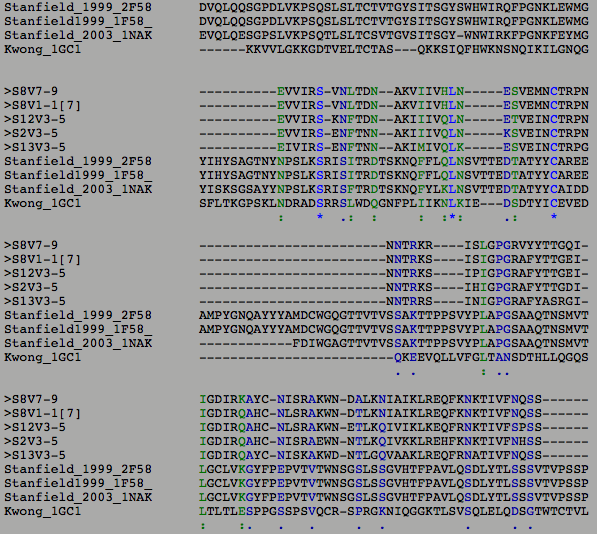
- The results of the Multiple Sequence Alignment can be seen below in the results.
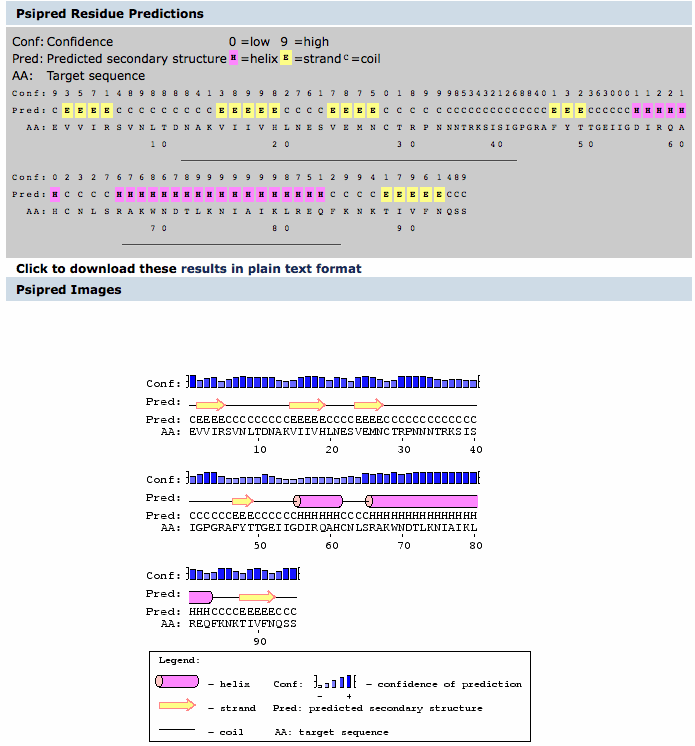
- After we were able to compare each of the amino acid sequences, we wanted to see what the secondary structure looked like.
- To do this, we used PSIPRED.
- We then used the 3D structures from the papers we presented during journal club.
- These structures were accessed through Cn3D.
- Since StarBiochem is easier for me to use, I imported the 1GC1, 1F58, 2F58, and 1NAK sequences from the paper onto this program, one at a time.
- The images below show the results I found when looking for the N and C termini, secondary structure elements, and the V3 region.
Results
- The green and black colors indicate areas where one or multiple subjects had an amino acid that diverged from what the others had.
- The image below is the secondary structure predicted using the PSIPRED program.
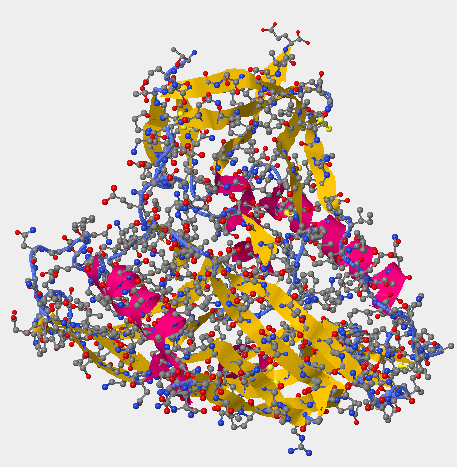
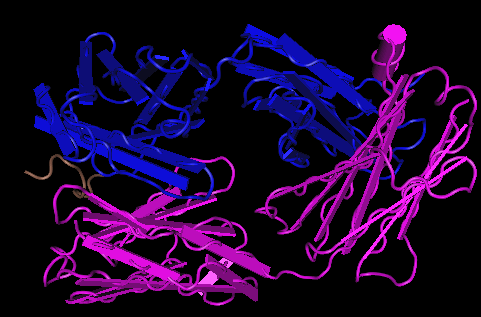
- The image below are the 3D structures generated from Kwong et al. (1998) structure 1GC1.
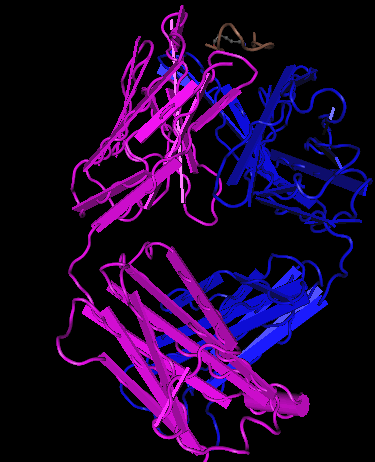
- The upper image on the right-hand side shows the secondary structures on gp 120 unlike the Cn3D image on the upper left which has the gp120, CD4, and the antibodies.
- The Kwong structure had no V3 region to identify.
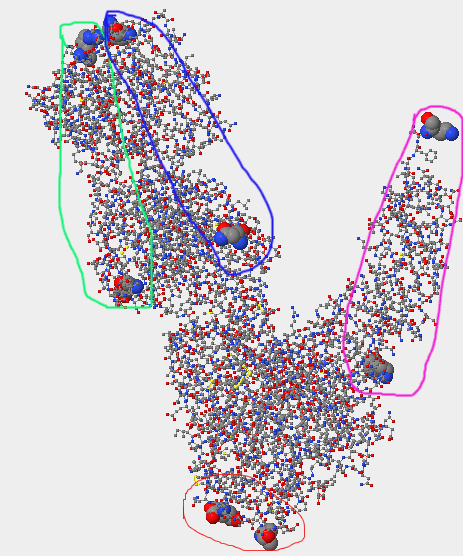
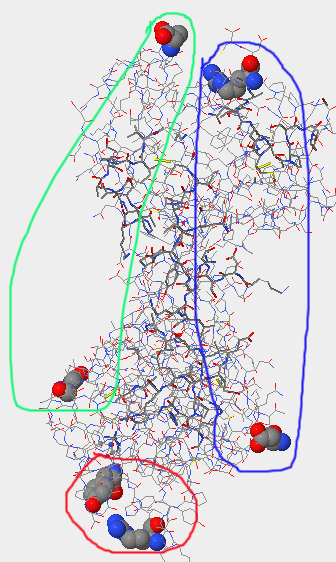
- The lower of the three images above is of the terminal ends; gp120 is circled in red, CD4 in pink, the antibody light-chain in light green, and the heavy chain in dark blue.
- For the 1GC1, the N-terminus for gp 120 is THR located at position 90. The C-terminus for gp 120 is GLU located at position 492. The N-terminus for the CD4 region is LYS at position 1, the C-terminus for CD4 is LYS at position 181. The N-terminus for the light chain of the antibody is GLU at position 1, the C-terminus for the light chain is ARG at position 213. The N-terminus for the heavy-chain is GLN at position 1, the C-terminus is LYS at position 229.
- The images below are the 3D structures generated from Stanfield et al. (1999) structure 1F58.
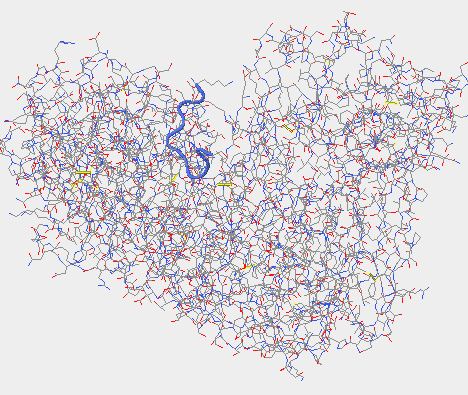
- The upper left-hand image was generated using Cn3D.
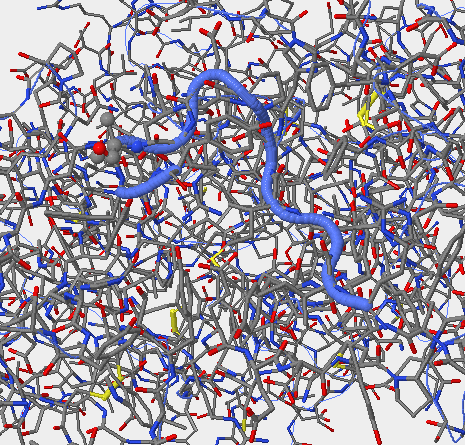
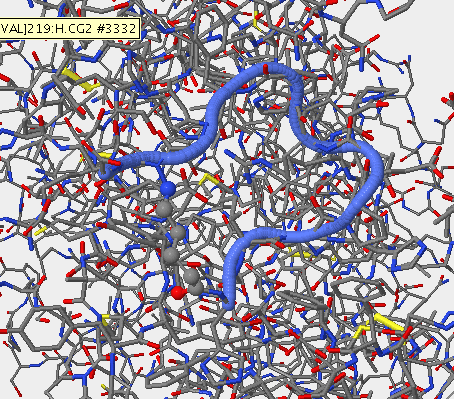
- The upper right-hand image was generated using Star Biochem, it shows the V3 region of gp 120 in it's secondary form which is a coil (blue).
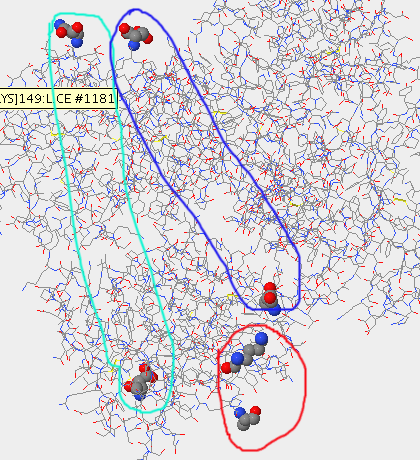
- The lower of the three images shows the terminal ends of the Stanfield 2F58 structure; the gp 120 is circle in red, the light chain of the antibody in light green and the heavy chain in dark blue.
- The N and C termini for gp 120 in this structure are ARG at position 313 and TYR at position 325, respectively. The N and C termini for the light chain are ASP at position 1 and ALA at position 212, respectively. The N and C termini for the heavy chain are ASP at position 1 and CYS at position 230, respectively. The N and C termini for the light chain are at ASP at position 1 and ALA at position 212, respectively.
- Below are the images of the 3D structures generated from Stanfield et al. (1999) structure 2F58.
- The upper left-hand image is the Cn3D image of the gp 120 and the antibodies.
- The upper right-hand image is the V3 region of the gp 120 in it's secondary structure which is a coil (blue).
- The lower of the three images shows the terminal ends of this structure; the gp 120 is circled in red, the light chain of the antibody is circled in light-green, and the heavy chain of the antibody is circled in dark blue.
- The N and C termini of the gp 120 are HIS at position 315 and PHE at position 324, respectively. The N and C termini for the heavy chain are ASP at position 1 and CYS at position 230, respectively. The N and C termini for the light chain are at ASP at position 1 and ALA at position 212, respectively.
- Below are the images of the 3D structure generated from Stanfield et al. (2003) structure 1NAK.
- The upper left-hand image shows the structure generated from Cn3D, that contains the gp 120 and the antibodies.
- The upper right-hand image shows the secondary structure of the v3 region on the gp 120 (the blue coil).
- The lower of the three images shows the terminal ends of the structure; gp 120 is circled in red, the light chain circled in light green, and the heavy chain in dark blue.
- The N and C termini of the gp 120 are LYS at position 312 and ALA at position 323, respectively. The N and C termini of the light chain are ASP at position 1 and ASN at position 212, respectively. The N and C termini of the heavy chain are GLU at position 1 and ASP at position 229, respectively.
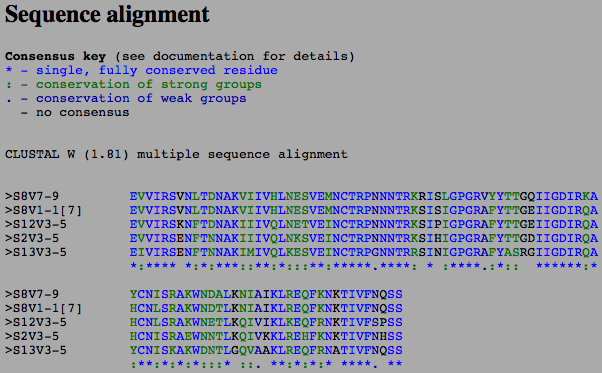
- The image below is the Multiple Sequence Alignment of just the subjects selected from the Markham study.
- There were some differences between subject 8 and the non-progressors that we think could account for the rate of progression of the virus. The secondary structure of the subjects were not affected by the changes in the amino acid sequences that the ClustalW tool displayed. This shows that this structure is important for the functioning of the virus and cannot be changed the way the virus constantly changes.
Links
BIOL368/F11:Class Journal Week 1
BIOL368/F11:Class Journal Week 2
BIOL368/F11:Class Journal Week 3
BIOL368/F11:Class Journal Week 4
BIOL368/F11:Class Journal Week 5
BIOL368/F11:Class Journal Week 6
BIOL368/F11:Class Journal Week 7
BIOL368/F11:Class Journal Week 8
BIOL368/F11:Class Journal Week 9
BIOL368/F11:Class Journal Week 10
BIOL368/F11:Class Journal Week 11
BIOL368/F11:Class Journal Week 12