Nicolette S. Harmon Week 14
DNA Microarray Project
Methods
- Files containing the microarray data from Berney et. al. 2010 paper were accessed via Array Express.
- The values from the various biological replicates in these files were merged into an excel spreadsheet and can be accessed at My Week 12 page.
- We had to normalize our data by multiplying by -1 for some of our values since the dyes were used in different orders in the Berney et. al. experiment.
- Once we normalized the data, we calculated the average, Tstat, and p values for each of the 3 experiments performed in the paper.
- The 2.5-50% Oxygen Saturation was the most significant, so that was the data we used for the rest of the analysis.
- GenMAPPFinder was then accessed to find the increasing and decreasing GO terms.
- The genes and their functions were determined below.
Files
Microarray Statistics Text File
MAPPFinder Results(Increased) Text File
MAPPFinder Results(Increased) SpreadSheet
MAPPFinder Results(Decreased) Text File
MAPPFinder Results(Decreased) SpreadSheet
Table of Significant Genes for 2.5-50 Saturation
p< Number of Genes 0.05 215 0.01 42 0.001 3
GO Terms
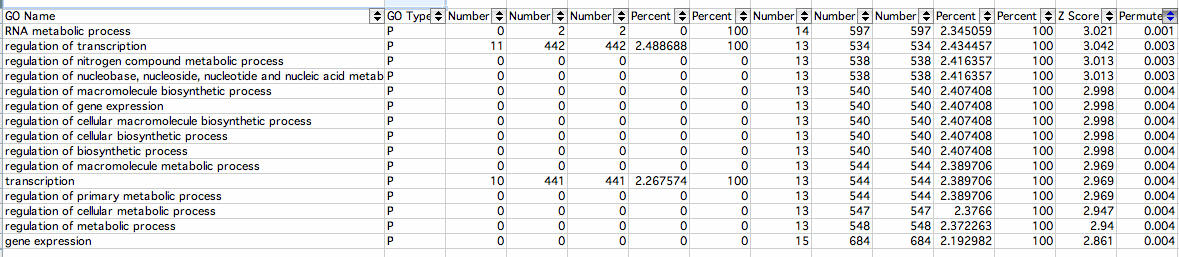
Increased:
- RNA Metabolic Process: The cellular chemical reactions and pathways involving RNA, ribonucleic acid, one of the two main type of nucleic acid, consisting of a long, unbranched macromolecule formed from ribonucleotides joined in 3',5'-phosphodiester linkage The Gene Ontology
- Regulation of Transcription: Any process that modulates the frequency, rate or extent of cellular DNA-dependent transcription The Gene Ontology
- Regulation of Nitrogen Compound Metabolic Process: Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving nitrogen or nitrogenous compounds The Gene Ontology
- Regulation of Nucleobase, Nucleoside, Nucleotide and Nucleic Acid Metabolic Process: Any cellular process that modulates the frequency, rate or extent of the chemical reactions and pathways involving nucleobases, nucleosides, nucleotides and nucleic acids The Gene Ontology
- Regulation of Macromolecule Biosynthetic Process: Any process that modulates the rate, frequency or extent of the chemical reactions and pathways resulting in the formation of a macromolecule. The Gene Ontology
- Regulation of Gene Expression: Any process that modulates the frequency, rate or extent of gene expression. This includes the production of an RNA transcript as well as any processing to produce a mature RNA product or an mRNA (for protein-coding genes) and the translation of that mRNA into protein The Gene Ontology
- Regulation of Cellular Macromolecule Biosynthetic Process: Any process that modulates the frequency, rate or extent of cellular macromolecule biosynthetic process The Gene Ontology
- Regulation of Cellular Biosynthetic Process: Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of substances, carried out by individual cells The Gene Ontology
- Regulation of Biosynthetic Process: Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of substances The Gene Ontology
- Regulation of Macromolecule Metabolic Process: Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving macromolecules. The Gene Ontology
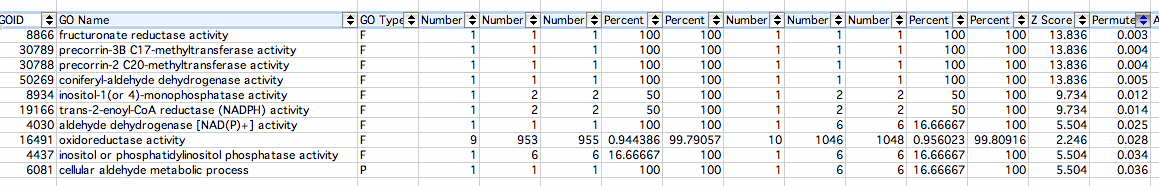
Decreased:
- Fructuronate Reductase Activity: Catalysis of the reaction: D-mannonate + NAD(+) = D-fructuronate + H(+) + NADH The Gene Ontology
- Precorrin-3B C17-methyltransferase Activity: Catalysis of the reaction: S-adenosyl-L-methionine + precorrin-3B = S-adenosyl-L-homocysteine + precorrin The Gene Ontology
- Precorrin-2 C20-methyltransferase Activity:Catalysis of the reaction: S-adenosyl-L-methionine + precorrin-2 = S-adenosyl-L-homocysteine + H(+) + precorrin-3A The Gene Ontology
- Coniferyl-aldehyde Dehydrogenase Activity: Catalysis of the reaction: coniferyl aldehyde + H2O + NAD(P)+ = ferulate + NAD(P)H + H+ The Gene Ontology
- Inositol-1(or 4)-monophosphatase Activity: Catalysis of the reaction: myo-inositol phosphate + H2O = myo-inositol + phosphate The Gene Ontology
- Trans-2-enoyl-CoA Reductase (NADPH) Activity: Catalysis of the reaction: acyl-CoA + NADP+ = trans-2,3-dehydroacyl-CoA + NADPH + H+ The Gene Ontology
- Aldehyde Dehydrogenase [NAD(P)+] Activity: Catalysis of the reaction: an aldehyde + NAD(P)+ + H2O = an acid + NAD(P)H + H+ The Gene Ontology
- Oxidoreductase Activity:The catalysis of redox reactions specifically relating to energy functions such as NAD+/NADH The Gene Ontology
- Inositol Phosphatase Activity: Catalysis of the reaction: inositol phosphate(n) + H2O = inositol phosphate(n-1) + phosphate. This reaction is the removal of a phosphate group from an inositol phosphate The Gene Ontology
- Cellular Aldehyde Metabolic Process: The chemical reactions and pathways involving aldehydes, any organic compound with the formula R-CH=O, as carried out by individual cells The Gene Ontology
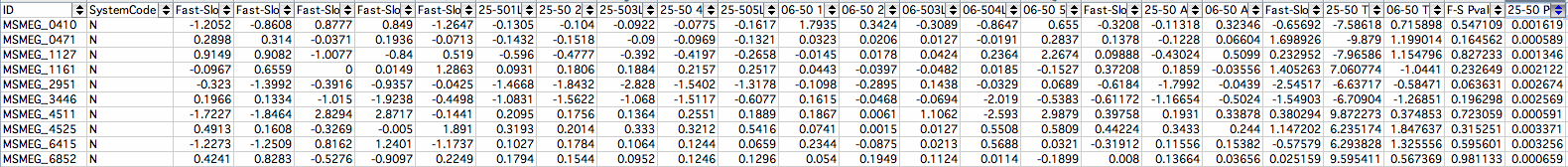
MSMEG IDs
- 5MSMEG_0410: MmpL Protein; P-Value: 0.001619 membrane protein involved in lipid transport
- 1MSMEG_0471: Transcriptional Regulator LysR Family Protein; P-Value: 0.000589
- 4MSMEG_1127: Probable Conserved Transmembrane Protein; P-Value: 0.001346
- 6MSMEG_1161: Taurine Transport System Permease Protein TauC; P-Value: 0.002122
- 8MSMEG_2951: [2Fe-2S] Binding Domain Protein; P-Value: 0.002674 Ferredoxin reducing/oxidizing enzyme powers TCA cycle independent of NAD/NADH
- 7MSMEG_3446: Hypothetical Protein; P-Value: 0.002569
- 2MSMEG_4511: Linear Gramicidin Synthetase mbtf; P-Value: 0.000591 antibiotic cells produces to kill other cells
- 10MSMEG_4525: Putative Oxygen-Independent Coproporhyrinogen III Oxidase; P-Value: 0.003371
- 9MSMEG_6415: Conserved Hypothetical Protein; P-Value: 0.003256
- 3MSMEG_6852: Putative Carboxylesterase/Lipase; P-Value: 0.000659 breaking down lipids
Discussion
Our statistical analysis correlated with the results found by Berney et. al. (2010). NAD+/NADH dependent enzymes were down-regulated while Ferredoxn and other regulators were up-regulated under these conditions. It is important for NAD+/NADH enzymes to be downregulated because they require more oxygen to run efficiently while Ferredoxn and other similar enzymes are considered "oxygen scanvengers" and can function properly in strenous conditions. When we ran GenMAPPFinder there were 221 errors that appeared according to the program so any descrepencies between our analysis and the paper is probably due to this.
Links
BIOL368/F11:Class Journal Week 1
BIOL368/F11:Class Journal Week 2
BIOL368/F11:Class Journal Week 3
BIOL368/F11:Class Journal Week 4
BIOL368/F11:Class Journal Week 5
BIOL368/F11:Class Journal Week 6
BIOL368/F11:Class Journal Week 7
BIOL368/F11:Class Journal Week 8
BIOL368/F11:Class Journal Week 9
BIOL368/F11:Class Journal Week 10
BIOL368/F11:Class Journal Week 11
BIOL368/F11:Class Journal Week 12