Nicolette S. Harmon Week 5
From OpenWetWare
Jump to navigationJump to search
Methods
- Subjects 3,7,8,11,12,13 were chosen based on what type of progressor they were and the number of sequences they had, they needed to have 50 or less in order to work on Workbench.
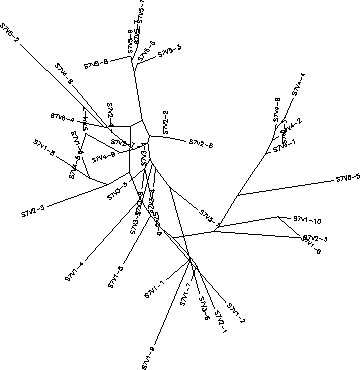
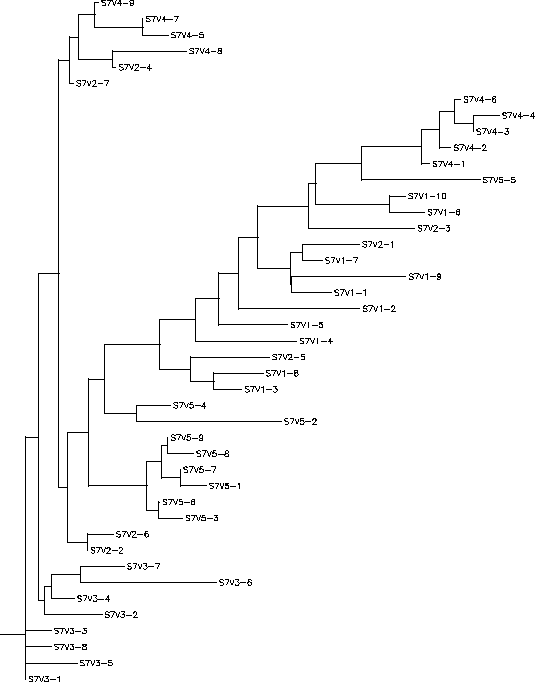
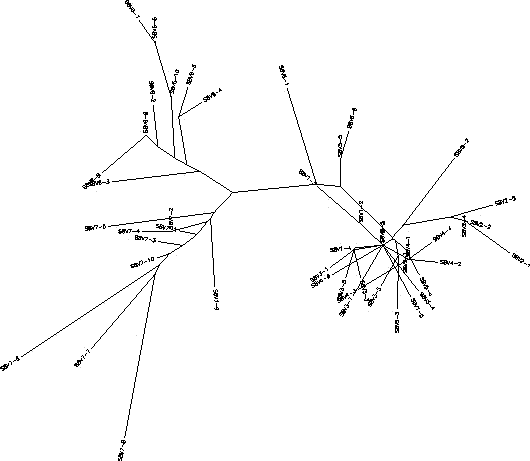
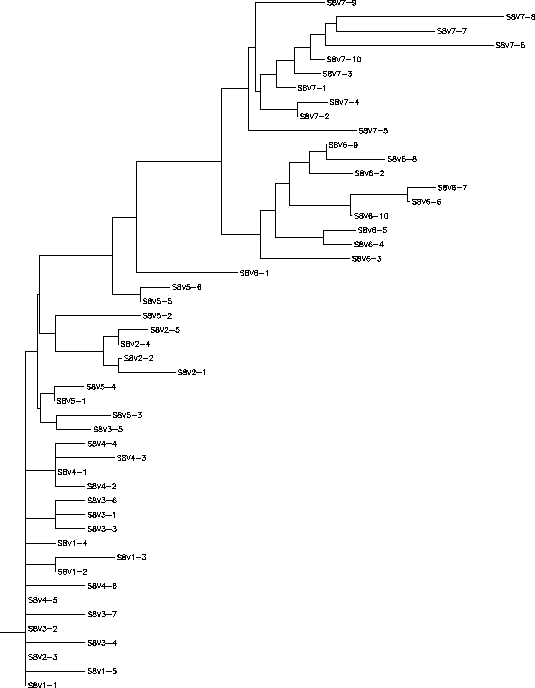
- I used the multiple alignment (ClustalW) tool on Workbench to make phylogenetic trees (both rooted and unrooted) for each subject.
- Both the unrooted and the rooted trees were uploaded in the results section below.
- Each subject's sequence alignment was imported so that the ClustalDist tool could be used.
- The ClustalDist tool measures the distance in divergence between any two sequences at a time, but only two per comparison.
- The minimum and maximum differences were calculated by multiplying the smallest/largest values by the total number of base pairs (this number was either 285bp or 288bp).
- The numbers for these values did not prove to be useful to my partner and I so we disregarded this data.
- Instead, we calculated the S value for each subject.
- This was done through the multiple alignment tool, where points of divergence in a given sequence were highlighted in black instead of the bright blue that the matching sequences were colored.
- Once we had our numbers for each subject, we realized we needed to find a way to normalize that data since there were inconsistencies between the subjects.
- The formula we used for normalizing our data was to divide the calculated value by the number of clones each subject had.
- Once we did that we had to divide these new values by the number of visits each subject came in for. This is important because this study was done over a span of four years and some subjects skipped multiple visits or they stopped coming in after a given number of times.
- Once we had our new S values we made a correlation between these numbers and the patterns we analyzed in the phylogenetic trees.
Results
Rapid Progressors
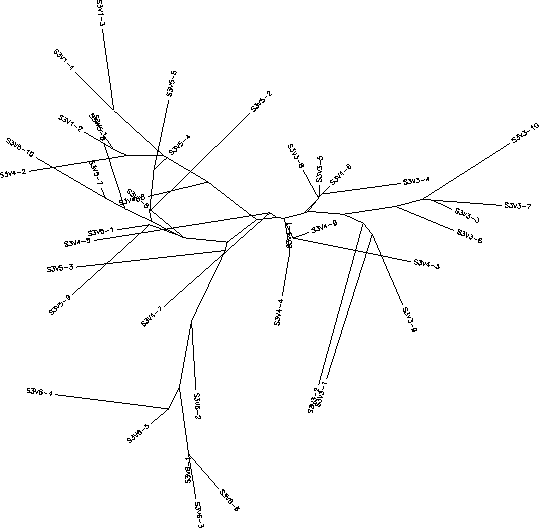
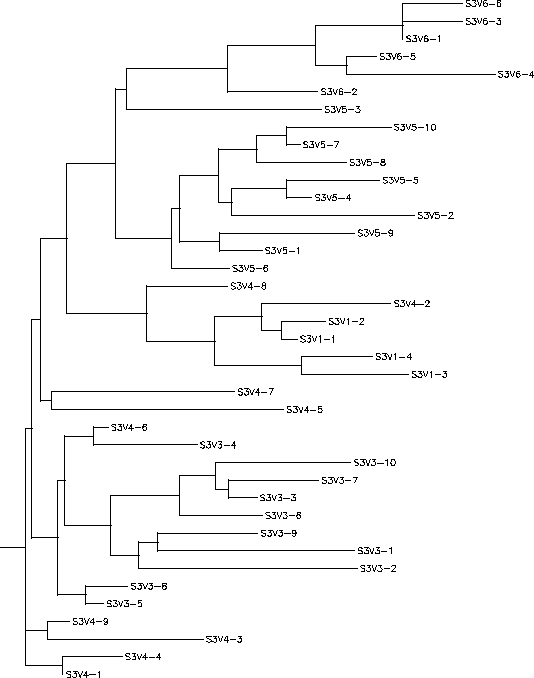
- The immune response in Subject 3 is selecting against a lack of change, meaning when the subject came in for each visit the virus had almost completely mutated into a new strain each time. Subject 3's immune response could be described as adaptive because it changed to correspond with the constant change HIV was undergoing. The rooted tree of Subject 3 shows that with each visit the virus had mutated it's sequence into something that was completely different from the previous visit, this information is indicated on the tree because all of the clones from a particular visit are in the same cluster which means they are fairly similar to each other. However, this constant change allowed for the immune system to ultimately not be able to keep up with this change which is why this subject's disease rapidly progressed. Subject 3 had a S value of S=42; the normalized value was 0.216.
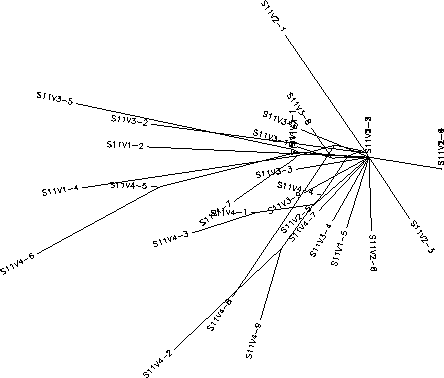
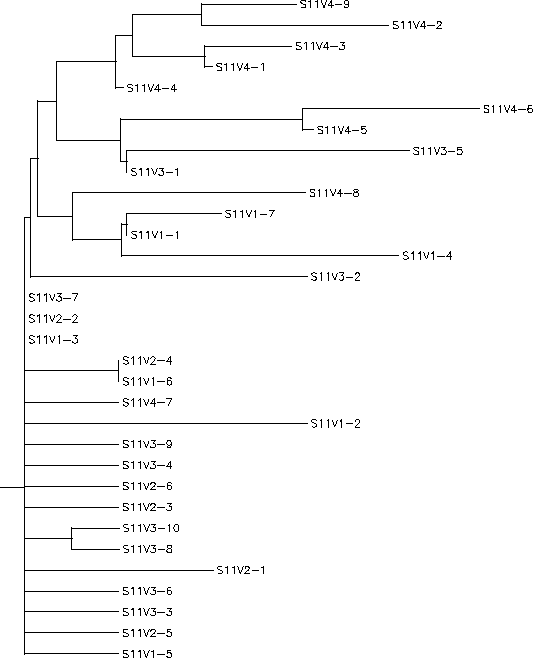
- Subject 11's immune system did not display any sort of selection (it was non-selective) in responding to the virus. There was a lot of diversity in between subject 11's visits meaning the clones in each visit were diverse, but they were similar to the clones from the past visits. Subject 11 only came in for 4 visits and by the fourth visit, the majority of clones were starting to become more diverse from the original pattern indicated by the first three visits. Had the subject come in for another visit it would have been interesting to see if the virus would continue to diverge from the previous sequences, if it would follow in similar sequence of visit 4 or if it would be like the first 3 visits. Subject 11 had a S value of S=34; the normalized value was 0.385.
Moderate Progressors
- The immune response to the virus in subject 7 indicated a "Best-Fit" pattern. When the subject came in for visit 3 the virus had mutated into a more diverse state than it previously was, however when the subject came in for the fourth visit this diversity had been completely knocked back by the immune system. The clones from visits 1,2,4 were all closely related and were therefore able to survive based on their sequences. The clones in visit 5 again mutated away from this successful sequence, however the subject did not come in for a sixth visit so it is unknown whether or not this mutation was selected against by the immune system. The S value for Subject 7 was S=54; the normalized value was 0.252.
- The clones in subject 8 started off not being very diverse in their nucleotide sequences for the first 5 visits. This would mean that the immune system was able to keep the virus in check by killing off equal numbers of each clone, we chose to call this a "Broad" response. However, when the subject came in for visits 6 and 7 there was a greater diversity amongst the clones that arose from a change in a few of the clones from visit 5. What should also be noted is that the clones from the sixth visit were different from the clones from the seventh visit indicating that the virus in this subject was mutating. The immune system was not able to keep up with this change in the virus and from there the virus grew more rapidly and became more diverse. Subject 8 starts out having a broad immune response to the virus, but over time the response becomes adaptive as the immune system is actively trying to keep up with the changing happening in the virus. The S value for Subject 8 was S=68; the normalized value was 0.199.
Non-Progressors
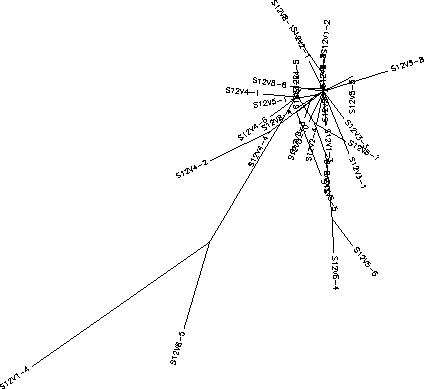
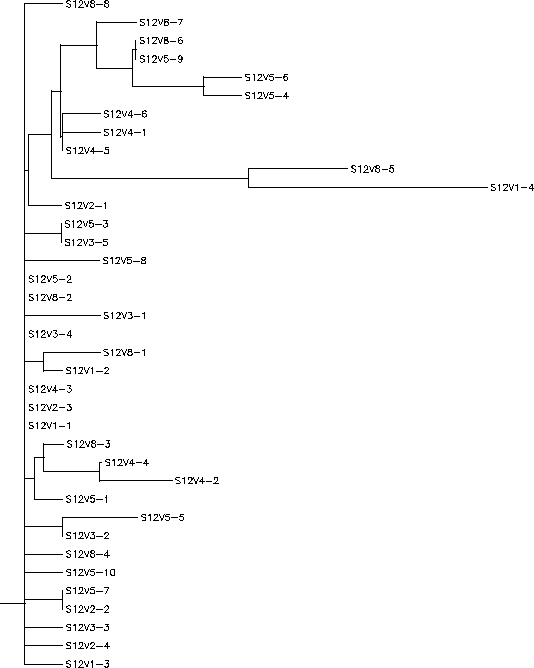
- Subject 12's immune system what we chose to call a "broad" response to the virus, it selected against change in the virus. Clone 4 in visit 1 diverged the most from the other strains, however it appeared that the immune system responded by knocking that particular strain back down. This kind of divergence did not reappear again until visit 8 when a clone emerged that was similar to this clone from visit 1. The clones from the other visits remain close to the"root" in the rooted tree, there is little divergence in these sequences. Subject 12 had a S value of S=42; and a normalized value of 0.191.
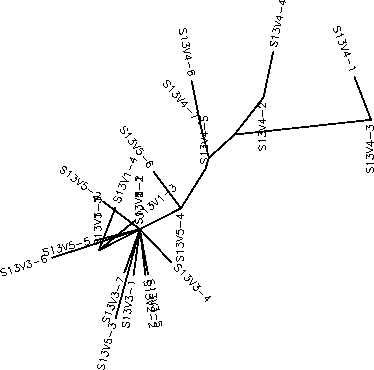
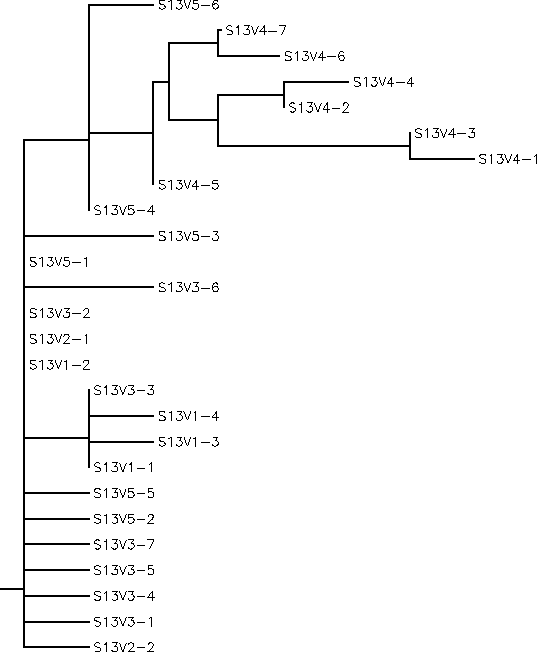
- The rooted tree for Subject 13 showed that the clones were closely related within each visit. Subject 13 had a similar immune response to that of Subject 12's, the broad response. However, Subject 13 happened to come in for a fourth visit when the virus had diverged from the sequence that was similar to the clones in the first 3 visits. When the Subject came back in for a fifth visit, none of the clones matched this mutation that the clones displayed in visit 4. It would appear that there was an "anti-evolution" because the clones in visit 5 reverted back to having similar sequences to those on the first three visits. This indicates that the immune system of this subject selected against change and if a change were to occur the immune system would wipe out the clones that mutated. Subject 13 had a S value of S=25; and a normalized value of 0.1924.
Reference Paper
Links
BIOL368/F11:Class Journal Week 1
BIOL368/F11:Class Journal Week 2
BIOL368/F11:Class Journal Week 3
BIOL368/F11:Class Journal Week 4
BIOL368/F11:Class Journal Week 5
BIOL368/F11:Class Journal Week 6
BIOL368/F11:Class Journal Week 7
BIOL368/F11:Class Journal Week 8
BIOL368/F11:Class Journal Week 9
BIOL368/F11:Class Journal Week 10
BIOL368/F11:Class Journal Week 11
BIOL368/F11:Class Journal Week 12