Zachary T. Goldstein Week 4
In Class HIV Activity
Purpose
The purpose of this lab is to practice analyzing HIV-1 nucleic sequence data using the Biology Workbench website so that we are better prepared for next week's activities. Sequences were uploaded, aligned, and compared by calculating "S" values, θ values, and min and max differences individually and between the subjects.
Methods/Results
Part I
- Uploaded the “visit_1_S1_S9.txt” and “visit_1_S10_S15.txt” files into the nucleic
acid tool set of the Biology Workbench by saving to ThawSpace
- Two data files were used because the Biology Workbench only allows up to 64
sequences to be uploaded at a time
- Generated a multiple sequence alignment and distance tree for 12 of these sequences (below)
- S15V1-7
- S15V1-8
- S15V1-9
- S10V1-6
- S10V1-5
- S10V1-4
- S1V1-2
- S1V1-3
- S1V1-4
- S6V1-1
- S6V1-2
- S6V1-3
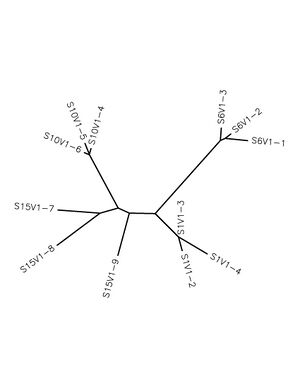
- Unrooted tree was created using Biology Workbench
- Selected sequences
- Selected "CLUSTALW" and then "Submit"
- The clones from each subject clustered together (Figure 1) however subject 15's sequences were more spread out than others
- Some subjects’ clones show more diversity than others. This is observable by tracing the length of lines connecting the sequences from the same subject (ex: S15 and S1) (Figure 1)
- None of the subjects really clustered together however it is difficult to differentiate the lengths of the terminal branches due to the lack of a definitive evolutionary beginning
- Observations of the tree:
- There is a clear close relationship between the sequences from the same subjects
- Sequences from subject 10 and 6 appear on opposite sides of the tree indicating a possible distant relationship
- There are otherwise no clear relationships between subjects' sequences due to no clear pattern of closeness of nodes
- The Unrooted Tree was saved and uploaded to this assignment (right)
Part II
- Selected all the clones from subjects 3, 14, and 1 respectively and ran separate alignments on set of sequences from each subject be selecting them and pressing "CLUSTALW" and "run"
- From the alignment 'S' values were calculated by counting the number of positions where there was at least one nucleotide difference across the collection of clones indicated by text not illuminated in blue
- Data was entered into Table 2 (below)
- Theta (θ) was calculated using formula provided in assignment and values were included in Table 2
- Theta calculations:
- Subject 3- 3.27
- Subject 14- 2.63
- Subject 1- 8.38
- Theta calculations:
- Min and Max differences were calculated via the Clustdist tool in the alignment tool set
- A distance matrix for each set of alignments (3) was completed and a table was produced
- The smallest and largest numbers were identified in the table and each were separately multiplied by 285 (representing the length of the sequence)
- This was done to convert that percentage difference score into the raw number of differences
- Results were rounded to the nearest integer (Table 2)
- Total sequences were compared to each other by importing sequences from 2 subjects and using the same procedure above (Table 3)
- 1 vs 3: min-0 max-41
- 1 vs 14: min-0 max-26
- 3 vs 14: min-1 max-42
| Subject | Number of Clones | "S" Value | Theta | Min Difference | Max Difference |
| 3 | 4 | 6 | 3.27 | 1 | 5 |
| 14 | 6 | 6 | 2.63 | 1 | 3 |
| 1 | 13 | 26 | 8.38 | 2 | 14 |
| Subjects Compared | Min Difference | Max Difference |
| 1 vs 3 | 0 | 41 |
| 1 vs 14 | 0 | 26 |
| 3 vs 14 | 1 | 42 |
Conclusion
The main purpose of this lab was to familiarize ourselves with Biology Workbench software and learn new ways to compare sequence data. Sequences were compared using "S" values, θ values, and by calculating min and max differences. This data allows us to understand and quantify some similarities and differences between sequences; calculations that can be used in later projects and assignments comparing data sets. Using min and max differences, it was interpreted that none of the three subjects were more closely related to any of the others; most min values were calculated to be 0 and most max values were quite large meaning there is high variation of the sequences between subjects.
Data and Files

Acknowledgments
- I received help on this assignment from User:Shivum A Desai
- While I received help on this page everything completed for this assignment was done by me and was not copied from anyone else
Zachary T. Goldstein 16:01, 22 September 2016 (EDT)Zachary T Goldstein
References
Markham, R.B., Wang, W.C., Weisstein, A.E., Wang, Z., Munoz, A., Templeton, A., Margolick, J., Vlahov, D., Quinn, T., Farzadegan, H., & Yu, X.F. (1998). Patterns of HIV-1 evolution in individuals with differing rates of CD4 T cell decline. Proc Natl Acad Sci U S A. 95, 12568-12573. doi: 10.1073/pnas.95.21.12568
Vlahov, D., Anthony, J.C., Munoz, A., Margolick, J., Nelson, K.E., Celentano, D.D., Solomon, L., Polk, B.F. (1991). The ALIVE study, a longitudinal study of HIV-1 infection in intravenous drug users: description of methods and characteristics of participants. NIDA Res Monogr 109, 75-100.
HIV Evolution Project
- What is your question?
- Does classification of subjects (3 groups) by dS/dN ratio instead of T cell count clarify the type of selection for or against certain types of mutations by the viral strains
- Make a prediction (hypothesis) about the answer to your question before you begin your analysis
- If ratios of dS/dN mutations are used as a means of comparison between groups (nonprogressor moderate progressor, and rapid progressor) it will provide evidence for selection for nonsynonymous mutations in viral strains observed in the progressor group because nonsynonymous mutations create diversity in the strain and make it harder for the immune system to fight it off. We also expect to see significantly larger average θ values for all subjects classified by the new rapid progressor group.
- Which subjects, visits, and clones will you use to answer your question
- For this experiment, 54 sequences were selected in total 18 from each of the three categories: rapid progressor, moderate progressor, and non progressor. This is to ensure that there is an adequate pool to compare the dS/dN ratios to see the differences between the three types of progressor groups. Thus, the following sequences are going to be used (accession numbers):
Rapid progressors
- AFO16760, AFO16761, AFO16762-subject 1 visit 1
- AFO16773, AFO16774, AFO16775-subject 1 visit 2
- AFO89109, AFO89110, AFO08911-subject 3 visit 1
- AFO89148, AFO89149, AFO89150-subject 4 visit 1
- AFO89151, AFO89152, AFO89153-subject 4 visit 2
- AFO89164, AFO89165, AFO89166-subject 4 visit 3
Moderate Progessors
- AF089195, AF089196, AFO89197-subject 5 visit 1
- AFO89203, AFO89204, AFO89205-subject 5 visit 2
- AFO89238, AFO89239, AFO89240-subject 6 visit 1
- AFO89241, AFO89242, AFO89243-subject 6 visit 2
- AFO89292, AFO89293, AFO89294-subject 7 visit 1
- AFO89335, AFO89336, AFO89337-subject 8 visit 1
Non-progessors
- AFO89529, AFO89530, AFO89531-subject 12 visit 1
- AFO89533, AFO89534, AFO89535-subject 12 visit 2
- AFO89566, AFO89567, AFO89568-subject 13 visit 1
- AFO89572, AFO89573, AFO89574-subject 13 visit 3
- AFO89579, AFO89580, AFO89581-subject 13 visit 4
- AFO89586, AFO89587, AFO89588-subject 13 visit 5
Acknowledgements
- I received help on this assignment from my partner User: Shivum A Desai in person and communicated with him via text message. Together we decided upon a question for the HIV Evolution project and randomly selected the subjects to use in our study
- I recieved help via email from User: Kam D. Dahlquist on formulating a more specific hypothesis
- Everything on this page was completed by myself and was not copied from anyone else
Zachary T. Goldstein 16:01, 22 September 2016 (EDT)Zachary T Goldstein
References
Markham, R.B., Wang, W.C., Weisstein, A.E., Wang, Z., Munoz, A., Templeton, A., Margolick, J., Vlahov, D., Quinn, T., Farzadegan, H., & Yu, X.F. (1998). Patterns of HIV-1 evolution in individuals with differing rates of CD4 T cell decline. Proc Natl Acad Sci U S A. 95, 12568-12573. doi: 10.1073/pnas.95.21.12568
Category: BIOL368/F16:People
BIOL368/F16
All class assignments:
- Week 1 Assignment
- Week 2 Assignment
- Week 3 Assignment
- Week 4 Assignment
- Week 5 Assignment
- Week 6 Assignment
- Week 7 Assignment
- Week 8 Assignment
- Week 9 Assignment
- Week 10 Assignment
- Week 11 Assignment
- Week 14 Assignment
- Week 15 Assignment
All individual assignments:
- Zachary T. Goldstein Week 2
- Zachary T. Goldstein Week 3
- Zachary T. Goldstein Week 4
- Zachary T. Goldstein Week 5
- Zachary T. Goldstein Week 6
- Zachary T. Goldstein Week 7
- Zachary T. Goldstein Week 8
- Zachary T. Goldstein Week 9
- Zachary T. Goldstein Week 10
- Zachary T. Goldstein Week 11
- Zachary T. Goldstein Week 14
- Zachary T. Goldstein Week 15
All shared journals:
- BIOL368/F16:Class Journal Week 1
- BIOL368/F16:Class Journal Week 2
- BIOL368/F16:Class Journal Week 3
- BIOL368/F16:Class Journal Week 4
- BIOL368/F16:Class Journal Week 5
- BIOL368/F16:Class Journal Week 6
- BIOL368/F16:Class Journal Week 7
- BIOL368/F16:Class Journal Week 8
- BIOL368/F16:Class Journal Week 9
- BIOL368/F16:Class Journal Week 10
- BIOL368/F16:Class Journal Week 11
- BIOL368/F16:Class Journal Week 14
- BIOL368/F16:Class Journal Week 15