Nida Patel Journal Week 4
Purpose
The purpose of this assignment is to familiarize ourselves with genetic code and phylogenetic trees to be able to assume relationships between species, thhis skill will be used in future analyses of data and genetic relations.
Methods/Results
GenBank
- I chose MK211376: Coronavirus BtRs-BetaCoVto analyze
- The accession number:YN2018B
- The page provides the complete genome of Coronavirus BtRs-BetaCoV, the source organism, the virus, the locus of the sequence, and referenced authors of the page.
- The assigned sequence for spike protein Bat SARS-like coronavirus RsSHC014 was saved onto a word processor and the spike protein sequence was as follows:
- >AGZ48806.1 spike protein [Bat SARS-like coronavirus RsSHC014]
MKLLVLVFATLVSSYTIEKCLDFDDRTPPANTQFLSSHRGVYYPDDIFRSNVLHLVQDHFLPFDSNVTRF ITFGLNFDNPIIPFRDGIYFAATEKSNVIRGWVFGSTMNNKSQSVIIMNNSTNLVIRACNFELCDNPFFV VLKSNNTQIPSYIFNNAFNCTFEYVSKDFNLDLGEKPGNFKDLREFVFRNKDGFLHVYSGYQPISAASGL PTGFNALKPIFKLPLGINITNFRTLLTAFPPRPDYWGTSAAAYFVGYLKPTTFMLKYDENGTITDAVDCS QNPLAELKCSVKSFEIDKGIYQTSNFRVAPSKEVVRFPNITNLCPFGEVFNATTFPSVYAWERKRISNCV ADYSVLYNSTSFSTFKCYGVSATKLNDLCFSNVYADSFVVKGDDVRQIAPGQTGVIADYNYKLPDDFLGC VLAWNTNSKDSSTSGNYNYLYRWVRRSKLNPYERDLSNDIYSPGGQSCSAVGPNCYNPLRPYGFFTTAGV GHQPYRVVVLSFELLNAPATVCGPKLSTDLIKNQCVNFNFNGLTGTGVLTPSSKRFQPFQQFGRDVSDFT DSVRDPKTSEILDISPCSFGGVSVITPGTNTSSEVAVLYQDVNCTDVPVAIHADQLTPSWRVYSTGNNVF QTQAGCLIGAEHVDTSYECDIPIGAGICASYHTVSSLRSTSQKSIVAYTMSLGADSSIAYSNNTIAIPTN FSISITTEVMPVSMAKTSVDCNMYICGDSTECANLLLQYGSFCTQLNRALSGIAVEQDRNTREVFAQVKQ MYKTPTLKDFGGFNFSQILPDPLKPTKRSFIEDLLFNKVTLADAGFMKQYGECLGDINARDLICAQKFNG LTVLPPLLTDDMIAAYTAALVSGTATAGWTFGAGAALQIPFAMQMAYRFNGIGVTQNVLYENQKQIANQF NKAISQIQESLTTTSTALGKLQDVVNQNAQALNTLVKQLSSNFGAISSVLNDILSRLDKVEAEVQIDRLI TGRLQSLQTYVTQQLIRAAEIRASANLAATKMSECVLGQSKRVDFCGKGYHLMSFPQAAPHGVVFLHVTY VPSQERNFTTAPAICHEGKAYFPREGVFVFNGTSWFITQRNFFSPQIITTDNTFVSGSCDVVIGIINNTV YDPLQPELDSFKEELDKYFKNHTSPDVDLGDISGINASVVNIQKEIDRLNEVAKNLNESLIDLQELGKYE QYIKWPWYVWLGFIAGLIAIVMVTILLCCMTSCCSCLKGACSCGSCCKFDEDDSEPVLKGVKLHYT
- The hyperlink was added to the data and tools section on the Week 4 Assignment Page and the spike sequence to the Week 4 Talk Page
Creating a phylogenetic tree with Phylogeny.fr
- I used www.phylogeny.frto do a phylogeny analysis on the sequences from the talk page.
- The steps were as listed:
- Click Phylogeny Analysis
- Click on One Click Mode
- Paste the talk page spike sequences
- Submit
- Click on tab 3.Alignment and under output reformat sequence into Alignment in Clustal Format
- The steps were as listed:
ALK02457.1 ---------MFIFLF------FLTLTSGSDLESCTT-------FDDVQAPNYPQHSSSRR
AAS10463.1 ---------MFIFLL------FLTLTSGSDLDRCTT-------FDDVQAPNYTQHTSSMR
AAP13441.1 ---------MFIFLL------FLTLTSGSDLDRCTT-------FDDVQAPNYTQHTSSMR
AAP13567.1 ---------MFIFLL------FLTLTSGSDLDRCTT-------FDDVQAPNYTQHTSSMR
AGZ48806.1 --------MKLLVLV------FATLVSSYTIEKCLD-------FDDRTPPANTQFLSSHR
QDF43825.1 --------MKLLVLV------FATLVSSYTIEKCTD-------FDDRTPPSNTQFLSSHR
AGZ48818.1 --------MKLLVLV------FATLVSSYTIEKCLD-------FDDRTPPANTQFLSSHR
QHD43416.1 ---------MFVFLV------LLPLVSS----QCVN-------LTTRTQLPPAYTNSFTR
AVP78031.1 ----------MLFFL------FLQFALVN--SQCVN-------LTGRTPLNPNYTNSSQR
ABD75323.1 --------MKILIFA------FL-VTLVKAQEGCGV-------INLRTQPKLTQVSSSRR
QDF43835.1 --------MKVLIVL------LC-LGLVTAQDGCGH-------ISTKPQPLLDKFSSSRR
ABD75332.1 --------MKVLIFA------LL-FSLAKAQEGCGI-------ISRKPQPKMEKVSSSRR
QDF43820.1 --------MKILIFA------FL-VTLVEAQEGCGI-------ISRKPQPKMAQVSSSRR
AAZ67052.1 --------MKILILA------FL-ASLAKAQEGCGI-------ISRKPQPKMAQVSSSRR
AFS88936.1 ----MIHSVFLLMFLLTPTESYVDVGPDSVKSACIEVDIQQT-FFDKTWPRPIDVSKAD-
YP_0010399 MTLLMCLLMSLLIFVRGCDSQFVDMSPASNTSECLESQVDAAAFSKLMWPYPIDPSKVD-
.:.. * : .
ALK02457.1 GVYYPDEIFRSDTLYLTQDLFLPFYSNVTGFHTINHR----------------FDNPVIP
AAS10463.1 GVYYPDEIFRSDTLYLTQDLFLPFYSNVTGFHTINHT----------------FDDPVIP
AAP13441.1 GVYYPDEIFRSDTLYLTQDLFLPFYSNVTGFHTINHT----------------FGNPVIP
AAP13567.1 GVYYPDEIFRSDTLYLTQDLFLPFYSNVTGFHTINHT----------------FDNPVIP
AGZ48806.1 GVYYPDDIFRSNVLHLVQDHFLPFDSNVTRFITFGLN----------------FDNPIIP
QDF43825.1 GVYYPDDIFRSNVLHLVQDHFLPFDSNVTRFITFGLN----------------FDNPIIP
AGZ48818.1 GVYYPDDIFRSNVLHLVQDHFLPFDSNVTRFITFGLN----------------FDNPIIP
QHD43416.1 GVYYPDKVFRSSVLHSTQDLFLPFFSNVTWFHAIHVSGTNGT---------KRFDNPVLP
AVP78031.1 GVYYPDTIYRSDTLVLSQGYFLPFYSNVSWYYSLTTN-NAAT---------KRTDNPILD
ABD75323.1 GVYYNDDIFRSDVLHLTQDYFLPFHSNLTQYFSLNIE-SDKI---------VYFDNPILK
QDF43835.1 GVYYNDDIFRSDVLHLTQDYFLPFDTNLTRYLSFNMD-SATK---------VYFDNPTLP
ABD75332.1 GVYYNDDIFRSDVLHLTQDYFLPFDSNLTQYFSLNID-SNKY---------TYFDNPILD
QDF43820.1 GVYYNDDIFRSDVLHLTQDYFLPFDSNLTQYFSLNVD-SDRY---------TYFDNPILD
AAZ67052.1 GVYYNDDIFRSNVLHLTQDYFLPFDSNLTQYFSLNVD-SDRF---------TYFDNPILD
AFS88936.1 GIIYPQGRTYSNITITYQGLF-PYQGDHGDMYVYSAGHATGT--TPQKLFVANYSQDVKQ
YP_0010399 GIIYPLGRTYSNITLAYTGLF-PLQGDLGSQYLYSVSHAVGHDGDPTKAYISNYSLLVND
*: * *. . * * : .
ALK02457.1 FKDGVYF----AATEKSNVVRG-------------WVFGSTMNNKSQS-VI--IINNSTN
AAS10463.1 FKDGIYF----AATEKSNVVRG-------------WVFGSTMNNKSQS-VI--IINNSTN
AAP13441.1 FKDGIYF----AATEKSNVVRG-------------WVFGSTMNNKSQS-VI--IINNSTN
AAP13567.1 FKDGIYF----AATEKSNVVRG-------------WVFGSTMNNKSQS-VI--IINNSTN
AGZ48806.1 FRDGIYF----AATEKSNVIRG-------------WVFGSTMNNKSQS-VI--IMNNSTN
QDF43825.1 FRDGVYF----AATEKSNVIRG-------------WVFGSTMNNKSQS-VI--IMNNSTN
AGZ48818.1 FKDGIYF----AATEKSNVIRG-------------WVFGSTMNNKSQS-VI--IMNNSTN
QHD43416.1 FNDGVYF----ASTEKSNIIRG-------------WIFGTTLDSKTQS-LL--IVNNATN
AVP78031.1 FKDGIYF----AATEHSNIIRG-------------WIFGTTLDNTSQS-LL--IVNNATN
ABD75323.1 FGDGVYF----AATEKSNVIRG-------------WVFGSTFDNTTQS-AI--IVNNSTH
QDF43835.1 FGDGIYF----AATEKSNVVRG-------------WIFGSTMDNTTQS-AI--IVNNSTH
ABD75332.1 FGDGVYF----AATEKSNVIRG-------------WIFGSSFDNTTQS-AI--IVNNSTH
QDF43820.1 FGDGVYF----AATEKSNVIRG-------------WIFGSTFDNTTQS-AV--IVNNSTH
AAZ67052.1 FGDGVYF----AATEKSNVIRG-------------WIFGSTFDNTTQS-AV--IVNNSTH
AFS88936.1 FANGFVVRIGAAANSTGTVIISPSTSATIRKIYPAFMLGSSVGNFSDG-KMGRFFNHTLV
YP_0010399 FDNGFVVRIGAAANSTGTIVISPSVNTKIKKAYPAFILGSSLTNTSAGQPL--YANYSLT
* :*. . *:.. ..:: . :::*::. . : . : * :
ALK02457.1 VVIRACNFELCDNPFFAVSKP-TGTQTHTM----IFDNAFNCTFEYISDS----FSLDVA
AAS10463.1 VVIRACNFELCDNPFFVVSKP-MGTRTHTM----IFDNAFNCTFEYISDA----FSLDVS
AAP13441.1 VVIRACNFELCDNPFFAVSKP-MGTQTHTM----IFDNAFNCTFEYISDA----FSLDVS
AAP13567.1 VVIRACNFELCDNPFFAVSKP-MGTQTHTM----IFDNAFNCTFEYISDA----FSLDVS
AGZ48806.1 LVIRACNFELCDNPFFVVLKS-NNTQIPSY----IFNNAFNCTFEYVSKD----FNLDLG
QDF43825.1 LVIRACNFELCDNPFFVVLRS-NNTQIPSY----IFNNAFNCTFEYVSKD----FNLDIG
AGZ48818.1 LVIRACNFELCDNPFFVVLKS-NNTQIPSY----IFNNAFNCTFEYVSKD----FNLDLG
QHD43416.1 VVIKVCEFQFCNDPFLGVYYH-KNNKSWMESEFRVYSSANNCTFEYVSQP----FLMDLE
AVP78031.1 VIIKVCNFDFCYDP-YLSGYY-HNNKTWSIREFAVYSSYANCTFEYVSKS----FMLNIS
ABD75323.1 IIIRVCYFNLCKDPMYTVSAG-TQKSSW------VYQSAFNCTYDRVEKS----FQLDTS
QDF43835.1 IIIRVCYFNLCKEPMYAISNE-QHYKSW------VYQNAYNCTYDRVEQS----FQLDTA
ABD75332.1 IIIRVCNFNLCKEPMYTVSKG-TQQSSW------VYQSAFNCTYDRVEKS----FQLDTA
QDF43820.1 IIIRVCNFNLCKEPMYTVSRG-TQQSSW------VYQSAFNCTYDRVERS----FQLDTA
AAZ67052.1 IIIRVCNFNLCKEPMYTVSRG-AQQSSW------VYQSAFNCTYDRVEKS----FQLDTA
AFS88936.1 LLPDGCGTLLR--AFYCILEPRSGNHCPAGNSYTSFATYHTPATDCSDGN----YNRNAS
YP_0010399 IIPDGCGTVLH--AFYCILKPRTVNRCPSGT---GYVSYF--IYETVHNDCQSTINRNAS
:: * : . : . : :
ALK02457.1 EKS-GNFKHLREFVFKNKDG--------FLYVYKGYQPIDVVRDLPSGF--NILKPIFKL
AAS10463.1 EKS-GNFKHLREFVFKNKDG--------FLYVYKGYQPIDVVRDLPSGF--NTLKPIFKL
AAP13441.1 EKS-GNFKHLREFVFKNKDG--------FLYVYKGYQPIDVVRDLPSGF--NTLKPIFKL
AAP13567.1 EKS-GNFKHLREFVFKNKDG--------FLYVYKGYQPIDVVRDLPSGF--NTLKPIFKL
AGZ48806.1 EKP-GNFKDLREFVFRNKDG--------FLHVYSGYQPISAASGLPTGF--NALKPIFKL
QDF43825.1 EKP-GNFKDLREFVFRNKDG--------FLHVYSGYQPISAASGLPTGF--NALKPIFKL
AGZ48818.1 EKP-GNFKDLREFVFRNKDG--------FLHVYSGYQPISAASGLPTGF--NALKPIFKL
QHD43416.1 GKQ-GNFKNLREFVFKNIDG--------YFKIYSKHTPINLVRDLPQGF--SALEPLVDL
AVP78031.1 GNG-GLFNTLREFVFRNVDG--------HFKIYSKFTPVNLNRGLPTGL--SVLQPLVEL
ABD75323.1 PKT-GNFTDLREFVFKNRDG--------FFTAYQTYTPVNLLRGLPSGL--SVLKPILKL
QDF43835.1 PQT-GNFKDLREYVFKNKDG--------FLSVYNAYSPIDIPRGLPVGF--SVLKPILKL
ABD75332.1 PKT-GNFKDLREYVFKNKGG--------FLRVYQTYTAVNLPRGFPAGF--SVLRPILKL
QDF43820.1 PKT-GNFKDLREYVFKNRDG--------FLSVYQTYTAVNLPRGLPIGF--SVLRPILKL
AAZ67052.1 PKT-GNFKDLREYVFKNRDG--------FLSVYQTYTAVNLPRGLPIGF--SVLRPILKL
AFS88936.1 LNSFKEYFNLRNCTFMYTYNITEDEILEWFGITQTAQGVHLFSSRYVDLYGGNMFQFATL
YP_0010399 LNSFKSFFDLVNCTFFNSWDITADETKEWFGITQDTQGVHLYSSRKGDLYGGNMFRFATL
: : * : .* . : . : . .: . : : *
ALK02457.1 PLGINITNFRAILTAF------LPAQDTWGTSAAAYFVGYLKPATFMLKYDENGTITDAV
AAS10463.1 PLGINITNFRAILTAF------SPAQDTWGTSAAAYFVGYLKPTTFMLKYDENGTITDAV
AAP13441.1 PLGINITNFRAILTAF------SPAQDIWGTSAAAYFVGYLKPTTFMLKYDENGTITDAV
AAP13567.1 PLGINITNFRAILTAF------SPAQDTWGTSAAAYFVGYLKPTTFMLKYDENGTITDAV
AGZ48806.1 PLGINITNFRTLLTAF------PPRPDYWGTSAAAYFVGYLKPTTFMLKYDENGTITDAV
QDF43825.1 PLGINITNFRTLLTAF------PPNPGYWGTSAAAYFVGYLKPTTFMLKYDENGTITDAV
AGZ48818.1 PLGINITNFRTLLTAF------PPRPDYWGTSAAAYFVGYLKPTTFMLKYDENGTITDAV
QHD43416.1 PIGINITRFQTLLALHRSYLTPGDSSSGWTAGAAAYYVGYLQPRTFLLKYNENGTITDAV
AVP78031.1 PVSINITKFRTLLTIHRGD---PMPNNGWTAFSAAYFVGYLKPRTFMLKYNENGTITDAV
ABD75323.1 PFGINITSFRVVMAMF------SKTTSNYVPESAAYYVGNLKQSTFMLSFNQNGTIVDAV
QDF43835.1 PIGINITSFKVVMSMF------SRTTSNFLPEVAAYFVGNLKYSTFMLNFNENGTITDAI
ABD75332.1 PFGINITSYRVVMTMF------SQFNSNFLPESAAYYVGNLKYTTFMLSFNENGTITDAV
QDF43820.1 PFGINITSYRVVMAMF------SQTTSNFLPESAAYYVGNLKYTTFMLRFNENGTITDAI
AAZ67052.1 PFGINITSYRVVMAMF------SQTTSNFLPESAAYYVGNLKYTTFMLSFNENGTITNAI
AFS88936.1 PVYDTIKYYSIIPHSIRSI---QSDRKAW----AAFYVYKLQPLTFLLDFSVDGYIRRAI
YP_0010399 PVYEGIKYYTVIPRSFRSK---ANKREAW----AAFYVYKLHQLTYLLDFSVDGYIRRAI
*. *. : : : **::* *: *::* :. :* * *:
ALK02457.1 DCSQNPLAELKCSVKSFEIDKGIYQTSNFRVAPSKEVVRFPNITNLCPFGEVFNATTFPS
AAS10463.1 DCSQNPLAELKCSVKSFEIDKGIYQTSNFRVVPSGDVVRFPNITNLCPFGEVFNATKFPS
AAP13441.1 DCSQNPLAELKCSVKSFEIDKGIYQTSNFRVVPSGDVVRFPNITNLCPFGEVFNATKFPS
AAP13567.1 DCSQNPLAELKCSVKSFEIDKGIYQTSNFRVVPSGDVVRFPNITNLCPFGEVFNATKFPS
AGZ48806.1 DCSQNPLAELKCSVKSFEIDKGIYQTSNFRVAPSKEVVRFPNITNLCPFGEVFNATTFPS
QDF43825.1 DCSQNPLAELKCSVKSFEIDKGIYQTSNFRVAPSKEVVRFPNITNLCPFGEVFNATTFPS
AGZ48818.1 DCSQNPLAELKCSVKSFEIDKGIYQTSNFRVAPSKEVVRFPNITNLCPFGEVFNATTFPS
QHD43416.1 DCALDPLSETKCTLKSFTVEKGIYQTSNFRVQPTESIVRFPNITNLCPFGEVFNATRFAS
AVP78031.1 DCALDPLSETKCTLKSLTVQKGIYQTSNFRVQPTQSVVRFPNITNVCPFHKVFNATRFPS
ABD75323.1 DCSQDPLAELKCTTKSFNVSKGIYQTSNFRVSPVTEVVRFPNITNLCPFDKVFNATRFPS
QDF43835.1 DCAQNPLSELKCTIKNFNVSKGIYQTSNFRVSPTHEVIRFPNITNRCPFDKVFNASRFPN
ABD75332.1 DCSQNPLAELKCTIKNFNVSKGIYQTSNFRVTPTQEVVRFPNITNRCPFDKVFNASRFPN
QDF43820.1 DCAQNPLAELKCTIKNFNVSKGIYQTSNFRVSPTQEVVRFPNITNRCPFDKVFNASRFPN
AAZ67052.1 DCAQNPLAELKCTIKNFNVSKGIYQTSNFRVSPTQEVIRFPNITNRCPFDKVFNATRFPN
AFS88936.1 DCGFNDLSQLHCSYESFDVESGVYSVSSFEAKPSGSVVEQAEGVE-CDFSPLLSGTP-PQ
YP_0010399 DCGHDDLSQLHCSYTSFEVDTGVYSVSSYEASATGTFIEQPNATE-CDFSPMLTGVA-PQ
**. : *:: :*: .: :..*:*..*.: . . .: .: .: * * ::.. ..
ALK02457.1 VYAWERKRISNCVADYSVLYNSTSFSTFKCYGVSATKLNDLCFSNVYADSFVVKGDDVRQ
AAS10463.1 VYAWERKRISNCVADYSVLYNSTSFSTFKCYGVSATKLNDLCFSNVYADSFVVKGDDVRQ
AAP13441.1 VYAWERKKISNCVADYSVLYNSTFFSTFKCYGVSATKLNDLCFSNVYADSFVVKGDDVRQ
AAP13567.1 VYAWERKKISNCVADYSVLYNSTFFSTFKCYGVSATKLNDLCFSNVYADSFVVKGDDVRQ
AGZ48806.1 VYAWERKRISNCVADYSVLYNSTSFSTFKCYGVSATKLNDLCFSNVYADSFVVKGDDVRQ
QDF43825.1 VYAWERKRISNCVADYSVLYNSTSFSTFKCYGVSATKLNDLCFSNVYADSFVVKGDDVRQ
AGZ48818.1 VYAWERKRISNCVADYSVLYNSTSFSTFKCYGVSATKLNDLCFSNVYADSFVVKGDDVRQ
QHD43416.1 VYAWNRKRISNCVADYSVLYNSASFSTFKCYGVSPTKLNDLCFTNVYADSFVIRGDEVRQ
AVP78031.1 VYAWERTKISDCIADYTVFYNSTSFSTFKCYGVSPSKLIDLCFTSVYADTFLIRFSEVRQ
ABD75323.1 VYAWERTKISDCVADYTVFYNSTSFSTFNCYGVSPSKLIDLCFTSVYADTFLIRFSEVRQ
QDF43835.1 VYAWERTKISDCVADYTVLYNSTSFSTFKCYGVSPSKLIDLCFTSVYADTFLIRSSEVRQ
ABD75332.1 VYAWERTKISDCVADYTVLYNSTSFSTFKCYGVSPSKLIDLCFTSVYADTFLIRSSEVRQ
QDF43820.1 VYAWERTKISDCVADYTVLYNSTSFSTFKCYGVSPSKLIDLCFTSVYADTFLIRSSEVRQ
AAZ67052.1 VYAWERTKISDCVADYTVLYNSTSFSTFKCYGVSPSKLIDLCFTSVYADTFLIRSSEVRQ
AFS88936.1 VYNFKRLVFTNCNYNLTKLLSLFSVNDFTCSQISPAAIASNCYSSLILDYFSYPLSMKSD
YP_0010399 VYNFKRLVFSNCNYNLTKLLSLFAVDEFSCNGISPDSIARGCYSTLTVDYFAYPLSMKSY
** ::* :::* : : : . .. *.* :*. : *::.: * * .
ALK02457.1 IAPGQTGVIADYNYKLPDDFTGC-VLAWNTRNIDATQTGNYNYKYRSLRHGKLRPFER-D
AAS10463.1 IAPGQTGVIADYNYKLPDDFMGC-VLAWNTRNIDATSTGNYNYKYRYLRHGKLRPFER-D
AAP13441.1 IAPGQTGVIADYNYKLPDDFMGC-VLAWNTRNIDATSTGNYNYKYRYLRHGKLRPFER-D
AAP13567.1 IAPGQTGVIADYNYKLPDDFMGC-VLAWNTRNIDATSTGNYNYKYRYLRHGKLRPFER-D
AGZ48806.1 IAPGQTGVIADYNYKLPDDFLGC-VLAWNTNSKDSSTSGNYNYLYRWVRRSKLNPYER-D
QDF43825.1 IAPGQTGVIADYNYKLPDDFMGC-VLAWNTRNIDATSTGNYNYKYRSLRHGKLRPFER-D
AGZ48818.1 IAPGQTGVIADYNYKLPDDFTGC-VLAWNTRNIDATQTGNYNYKYRSLRHGKLRPFER-D
QHD43416.1 IAPGQTGKIADYNYKLPDDFTGC-VIAWNSNNLDSKVGGNYNYLYRLFRKSNLKPFER-D
AVP78031.1 VAPGQTGVIADYNYKLPDDFTGC-VIAWNTAKQD---VGNYF--YRSHRSTKLKPFER-D
ABD75323.1 VAPGQTGVIADYNYKLPDDFTGC-VIAWNTAKQD---VGSYF--YRSHRSSKLKPFER-D
QDF43835.1 VAPGETGVIADYNYKLPDDFTGC-VIAWNTAKQD---QGQYY--YRSSRKTKLKPFER-D
ABD75332.1 VAPGETGVIADYNYKLPDDFTGC-VIAWNTAQQD---QGQYY--YRSYRKEKLKPFER-D
QDF43820.1 VAPGETGVIADYNYKLPDDFTGC-VIAWNTAKQD---TGHYY--YRSHRKTKLKPFER-D
AAZ67052.1 VAPGETGVIADYNYKLPDDFTGC-VIAWNTAKQD---QGQYY--YRSHRKTKLKPFER-D
AFS88936.1 LSVSSAGPISQFNYKQSFSNPTC-LILATVPHNLTTITKPLKYSYIN-KCSRLLSDDRTE
YP_0010399 IRPGSAGNIPLYNYKQSFANPTCRVMASVLANVTITKPHAYG--YIS-KCSRLTGANQ-D
: ..:* *. :*** . * :: * . .* :. :
ALK02457.1 ISNVPFSPDGK--PCTPP-AFNCYW-----------PLNDYGFYITNGIGYQPYRVVVLS
AAS10463.1 ISNVPFSPDGK--PCTPP-APNCYW-----------PLNGYGFYTTSGIGYQPYRVVVLS
AAP13441.1 ISNVPFSPDGK--PCTPP-ALNCYW-----------PLNDYGFYTTTGIGYQPYRVVVLS
AAP13567.1 ISNVPFSPDGK--PCTPP-ALNCYW-----------PLNDYGFYTTTGIGYQPYRVVVLS
AGZ48806.1 LSNDIYSPGGQ--SCSAV-GPNCYN-----------PLRPYGFFTTAGVGHQPYRVVVLS
QDF43825.1 ISNVPFSPDGK--PCTPP-AFNCYW-----------PLNDYGFFTTNGIGYQPYRVVVLS
AGZ48818.1 ISNVPFSPDGK--PCTPP-AFNCYW-----------PLNDYGFYITNGIGYQPYRVVVLS
QHD43416.1 ISTEIYQAGST--PCNGVEGFNCYF-----------PLQSYGFQPTNGVGYQPYRVVVLS
AVP78031.1 LSS----------------DENGVR-----------TLSTYDFNPNVPLEYQATRVVVLS
ABD75323.1 LSS----------------EENGVR-----------TLSTYDFNQNVPLEYQATRVVVLS
QDF43835.1 LTS----------------DENGVR-----------TLSTYDFYPNVPIEYQATRVVVLS
ABD75332.1 LSS----------------DENGVY-----------TLSTYDFYPSIPVEYQATRVVVLS
QDF43820.1 LSSD---------------DGNGVY-----------TLSTYDFNPNVPVAYQATRVVVLS
AAZ67052.1 LSS----------------DENGVR-----------TLSTYDFYPSVPVAYQATRVVVLS
AFS88936.1 VPQLVNANQYS--PCVSI-VPSTVWEDGDYYRKQLSPLEGGGWLVASGSTVAMTEQLQMG
YP_0010399 VETPLYINPGEYSICRDF-SPGGFSEDGQVFKRTLTQFEGGGLLIGVGTRVPMTDNLQMS
: . : . : :.
ALK02457.1 FELL----NAPATVC-----GPKLSTDLIKNQCVNFNFNGLTGTGVLTPSSKRFQPFQQF
AAS10463.1 FELL----NAPATVC-----GPKLSTDLIKNQCVNFNFNGLTGTGVLTPSSKRFQPFQQF
AAP13441.1 FELL----NAPATVC-----GPKLSTDLIKNQCVNFNFNGLTGTGVLTPSSKRFQPFQQF
AAP13567.1 FELL----NAPATVC-----GPKLSTDLIKNQCVNFNFNGLTGTGVLTPSSKRFQPFQQF
AGZ48806.1 FELL----NAPATVC-----GPKLSTDLIKNQCVNFNFNGLTGTGVLTPSSKRFQPFQQF
QDF43825.1 FELL----NAPATVC-----GPKLSTDLIKNQCVNFNFNGLTGTGVLTPSSKRFQPFQQF
AGZ48818.1 FELL----NAPATVC-----GPKLSTDLIKNQCVNFNFNGLTGTGVLTPSSKRFQPFQQF
QHD43416.1 FELL----HAPATVC-----GPKKSTNLVKNKCVNFNFNGLTGTGVLTESNKKFLPFQQF
AVP78031.1 FELL----NAPATVC-----GPKLSTQLVKNQCVNFNFNGLKGTGVLTDSSKRFQSFQQF
ABD75323.1 FELL----NAPATVC-----GPKLSTSLVKNQCVNFNFNGFKGTGVLTDSSKTFQSFQQF
QDF43835.1 FELL----NAPATVC-----GPKLSTGLVKNQCVNFNFNGLRGTGVLTDSSKRFQSFQQF
ABD75332.1 FELL----NAPATVC-----GPKLSTQLVKNQCVNFNFNGLRGTGVLTTSSKRFQSFQQF
QDF43820.1 FELL----NAPATVC-----GPKLSTQLVKNQCVNFNFNGLKGTGVLTDSSKRFQSFQQF
AAZ67052.1 FELL----NAPATVC-----GPKLSTQLVKNQCVNFNFNGLKGTGVLTESSKRFQSFQQF
AFS88936.1 FGITVQYGTDTNSVCPKLEFANDTKIASQLGNCVEYSLYGVSGRGVFQNCTAVGVRQQRF
YP_0010399 FIISVQYGTGTDSVCPMLDLGDSLTITNRLGKCVDYSLYGVTGRGVFQNCTAVGVKQQRF
* : . :** . . . .:**::.: *. * **: .. *.*
ALK02457.1 GRDVLD-FTDSVRDPKTSEILDISPCSFGGVSVITPGTNTSSEVAVLYQDVNCTDVPVAI
AAS10463.1 GRDVSD-FTDSVRDPKTSEILDISPCSFGGVSVITPGTNASSEVAVLYQDVNCTDVSTLI
AAP13441.1 GRDVSD-FTDSVRDPKTSEILDISPCSFGGVSVITPGTNASSEVAVLYQDVNCTDVSTAI
AAP13567.1 GRDVSD-FTDSVRDPKTSEILDISPCSFGGVSVITPGTNASSEVAVLYQDVNCTDVSTAI
AGZ48806.1 GRDVSD-FTDSVRDPKTSEILDISPCSFGGVSVITPGTNTSSEVAVLYQDVNCTDVPVAI
QDF43825.1 GRDVSD-FTDSVRDPKTSEILDISPCSFGGVSVITPGTNTSSEVAVLYQDVNCTDVPVAI
AGZ48818.1 GRDVSD-FTDSVRDPKTSEILDISPCSFGGVSVITPGTNTSSEVAVLYQDVNCTDVPVAI
QHD43416.1 GRDIAD-TTDAVRDPQTLEILDITPCSFGGVSVITPGTNTSNQVAVLYQDVNCTEVPVAI
AVP78031.1 GKDASD-FIDSVRDPQTLEILDITPCSFGGVSVITPGTNTSLEVAVLYQDVNCTDVPTTI
ABD75323.1 GRDASD-FTDSVRDPQTLRILDISPCSFGGVSVITPGTNTSSAVAVLYQDVNCTDVPRTI
QDF43835.1 GRDTSD-FTDSVRDPQTLEILDITPCSFGGVSVITPGTNASSEVAVLYQDVNCTDVPTAI
ABD75332.1 GRDTSD-FTDSVRDPQTLEILDISPCSFGGVSVITPGTNASSEVAVLYQDVNCTDVPTSI
QDF43820.1 GRDTSD-FTDSVRDPQTLEILDITPCSFGGVSVITPGTNASSEVAVLYQDVNCTDVPTAI
AAZ67052.1 GRDTSD-FTDSVRDPQTLEILDISPCSFGGVSVITPGTNASSEVAVLYQDVNCTDVPAAI
AFS88936.1 VYDAYQNLVGYYSDDGNYYCLR--ACVSVPVSVIY--DKETKTHATLFGSVACEHISSTM
YP_0010399 VYDSFDNLVGYYSDDGNYYCVR--PCVSVPVSVIY--DKSTNLHATLFGSVACEHVTTMM
* : . * . : .* **** : : *.*: .* * :. :
ALK02457.1 -HADQLTPS-WRVYSTGNNVFQTQAGCLIGAEHVDTSY---ECDIPIGAGICASYHTVSS
AAS10463.1 -HAEQLTPA-WRIYSTGNNVFQTQAGCLIGAEHVDTSY---ECDIPIGAGICASYHTVSS
AAP13441.1 -HADQLTPA-WRIYSTGNNVFQTQAGCLIGAEHVDTSY---ECDIPIGAGICASYHTVSL
AAP13567.1 -HADQLTPA-WRIYSTGNNVFQTQAGCLIGAEHVDTSY---ECDIPIGAGICASYHTVSL
AGZ48806.1 -HADQLTPS-WRVYSTGNNVFQTQAGCLIGAEHVDTSY---ECDIPIGAGICASYHTVSS
QDF43825.1 -HADQLTPA-WRIYSTGNNVFQTQAGCLIGAEHVDTSY---ECDIPIGAGICASYHTVSS
AGZ48818.1 -HADQLTPS-WRVYSTGNNVFQTQAGCLIGAEHVDTSY---ECDIPIGAGICASYHTVSS
QHD43416.1 -HADQLTPT-WRVYSTGSNVFQTRAGCLIGAEHVNNSY---ECDIPIGAGICASYQTQTN
AVP78031.1 -HADQLTPA-WRIYATGTNVFQTQAGCLIGAEHVNASY---ECDIPIGAGICASYHTASI
ABD75323.1 -QADQLAPS-WRVYTTGPYVFQTQAGCLIGAEHVNASY---QCDIPIGAGICASYHTASH
QDF43835.1 -RADQLTPA-WRVYSTGINVFQTQAGCLIGAEHVNASY---ECDIPIGAGICASYHTAST
ABD75332.1 -HADQLTPA-WRVYSTGVNVFQTQAGCLIGAEHVNASY---ECDIPIGAGICASYHTASV
QDF43820.1 -RADQLTPA-WRVYSTGVNVFQTQAGCLIGAEHVNASY---ECDIPIGAGICASYHTAST
AAZ67052.1 -HADQLTPA-WRVYSTGTNVFQTQAGCLIGAEHVNASY---ECDIPIGAGICASYHTAST
AFS88936.1 SQYSRSTRSMLKRRDSTYGPLQTPVGCVLGL--VNSSLFVEDCKLPLGQSLCALPDTPST
YP_0010399 SQFSRLTQS-NLRRRDSNIPLQTAVGCVIGLS--NNSLVVSDCKLPLGQSLCAVPP-VST
. .. : : :** .**::* : * :*.:*:* .:** :
ALK02457.1 ----LRSTS----QKSI--------VAYTMSLGADSSIAYSNNTIAIPTNFSISITTEVM
AAS10463.1 ----LRSTS----QKSI--------VAYTMSLGADSSIAYSNNTIAIPTNFSISITTEVM
AAP13441.1 ----LRSTS----QKSI--------VAYTMSLGADSSIAYSNNTIAIPTNFSISITTEVM
AAP13567.1 ----LRSTS----QKSI--------VAYTMSLGADSSIAYSNNTIAIPTNFSISITTEVM
AGZ48806.1 ----LRSTS----QKSI--------VAYTMSLGADSSIAYSNNTIAIPTNFSISITTEVM
QDF43825.1 ----LRSTS----QKSI--------VAYTMSLGADSSIAYSNNTIAIPTNFSISITTEVM
AGZ48818.1 ----LRSTS----QKSI--------VAYTMSLGADSSIAYSNNTIAIPTNFSISITTEVM
QHD43416.1 SPRRARSVA----SQSI--------IAYTMSLGAENSVAYSNNSIAIPTNFTISVTTEIL
AVP78031.1 ----LRSTS----QKAI--------VAYTMSLGAENSIAYANNSIAIPTNFSISVTTEVM
ABD75323.1 ----LRSTG----QKSI--------VAYTMSLGAENSVAYANNSIAIPTNFSISVTTEVM
QDF43835.1 ----LRSVG----QKSI--------VAYTMSLGAENSIAYANNSIAIPTNFSISVTTEVM
ABD75332.1 ----LRSTG----QKSI--------VAYTMSLGAENSIAYANNSIAIPTNFSISVTTEVM
QDF43820.1 ----LRSVG----QKSI--------VAYTMSLGAENSIAYANNSIAIPTNFSISVTTEVM
AAZ67052.1 ----LRSVG----QKSI--------VAYTMSLGAENSIAYANNSIAIPTNFSISVTTEVM
AFS88936.1 ----LTPRS----VRSVPGEMRLASIAFNHPIQVDQ-LNSSYFKLSIPTNFSFGVTQEYI
YP_0010399 ----FRSYSASQFQLAV--------LNYTSPI-VVTPINSSGFTAAIPTNFSFSVTQEYI
. . :: : :. .: . : : . :*****::.:* * :
ALK02457.1 PVSMAKTSVDCNMYICGDSTECANLLLQYGSFCTQLNRALSGIAVEQDRNTREVFAQVKQ
AAS10463.1 PVSMAKTSVDCNMYICGDSTECANLLLQYGSFCRQLNRALSGIAAEQDRNTREVFVQVKQ
AAP13441.1 PVSMAKTSVDCNMYICGDSTECANLLLQYGSFCTQLNRALSGIAAEQDRNTREVFAQVKQ
AAP13567.1 PVSMAKTSVDCNMYICGDSTECANLLLQYGSFCTQLNRALSGIAAEQDRNTREVFAQVKQ
AGZ48806.1 PVSMAKTSVDCNMYICGDSTECANLLLQYGSFCTQLNRALSGIAVEQDRNTREVFAQVKQ
QDF43825.1 PVSMAKTSVDCNMYICGDSTECANLLLQYGSFCTQLNRALSGIAVEQDRNTREVFAQVKQ
AGZ48818.1 PVSMAKTSVDCNMYICGDSTECANLLLQYGSFCTQLNRALSGIAVEQDRNTREVFAQVKQ
QHD43416.1 PVSMTKTSVDCTMYICGDSTECSNLLLQYGSFCTQLNRALTGIAVEQDKNTQEVFAQVKQ
AVP78031.1 PVSMAKTSVDCTMYICGDSIECSNLLLQYGSFCTQLNRALSGIAIEQDKNTQEVFAQVKQ
ABD75323.1 PVSMAKTSVDCTMYICGDSLECSNLLLQYGSFCTQLNRALSGIAVEQDKNTQEVFAQVKQ
QDF43835.1 PVSMSKTSVDCTMYICGDSQECSNLLLQYGSFCTQLNRALTGIAIEQDKNTQEVFAQVKQ
ABD75332.1 PVSIAKTSVDCTMYICGDSLECSNLLLQYGSFCTQLNRALTGIAIEQDKNTQEVFAQVKQ
QDF43820.1 PVSMAKTSVDCTMYICGDSQECSNLLLQYGSFCTQLNRALTGVALEQDKNTQEVFAQVKQ
AAZ67052.1 PVSMAKTSVDCTMYICGDSLECSNLLLQYGSFCTQLNRALSGIAIEQDKNTQEVFAQVKQ
AFS88936.1 QTTIQKVTVDCKQYVCNGFQKCEQLLREYGQFCSKINQALHGANLRQDDSVRNLFASVKS
YP_0010399 ETSIQKVTVDCKQYVCNGFTRCEKLLVEYGQFCSKINQALHGANLRQDESVYSLYSNIKT
.:: *.:***. *:*.. * :** :**.** ::*.** * ** .. .:: .:*
ALK02457.1 MYKTPTLKDFGG-FNFSQILPDPLKPTKRSF---IEDLLFNKVTLADAGFMKQYGECL--
AAS10463.1 MYKTPTLKDFGG-FNFSQILPDPLKPTKRSF---IEDLLFNKVTLADAGFMKQYGECL--
AAP13441.1 MYKTPTLKYFGG-FNFSQILPDPLKPTKRSF---IEDLLFNKVTLADAGFMKQYGECL--
AAP13567.1 MYKTPTLKYFGG-FNFSQILPDPLKPTKRSF---IEDLLFNKVTLADAGFMKQYGECL--
AGZ48806.1 MYKTPTLKDFGG-FNFSQILPDPLKPTKRSF---IEDLLFNKVTLADAGFMKQYGECL--
QDF43825.1 MYKTPTLKDFGG-FNFSQILPDPLKPTKRSF---IEDLLFNKVTLADAGFMKQYGECL--
AGZ48818.1 MYKTPTLKDFGG-FNFSQILPDPLKPTKRSF---IEDLLFNKVTLADAGFMKQYGECL--
QHD43416.1 IYKTPPIKDFGG-FNFSQILPDPSKPSKRSF---IEDLLFNKVTLADAGFIKQYGDCL--
AVP78031.1 IYKTPPIKDFGG-FNFSQILPDPSKPSKRSF---IEDLLFNKVTLADAGFIKQYGDCL--
ABD75323.1 MYKTPTIRDFGG-FNFSQILPDPLKPTKRSF---IEDLLYNKVTLADAGFMKQYADCL--
QDF43835.1 MYKTPAIKDFGG-FNFSQILPDPSKPTKRSF---IEDLLFNKVTLADAGFMKQYGECL--
ABD75332.1 MYKTPAIKDFGG-FNFSQILPDPSKPTKRSF---IEDLLFNKVTLADAGFMKQYGECL--
QDF43820.1 MYKTPAIKDFGG-FNFSQILPDPSKPTKRSF---IEDLLFNKVTLADAGFMKQYGECL--
AAZ67052.1 MYKTPAIKDFGG-FNFSQILPDPSKPTKRSF---IEDLLFNKVTLADAGFMKQYGECL--
AFS88936.1 SQSSPIIPGFGGDFNLTLLEPVSISTGSRSARSAIEDLLFDKVTIADPGYMQGYDDCMQQ
YP_0010399 TSTQTLEYGLNGDFNLTLLQVPQIGGSSSSYRSAIEDLLFDKVTIADPGYMQGYDDCMKQ
. . :.* **:: : . * *****::***:**.*::: * :*:
ALK02457.1 GDINARDLICAQKFNGLTVLPPLLTDDMIAAYTAALVSGTATAGWTFGAGAALQIPFAMQ
AAS10463.1 GDINARDLICAQKFNGLTVLPPLLTDDMIAAYTAALVSGTATAGWTFGAGAALQIPFAMQ
AAP13441.1 GDINARDLICAQKFNGLTVLPPLLTDDMIAAYTAALVSGTATAGWTFGAGAALQIPFAMQ
AAP13567.1 GDINARDLICAQKFNGLTVLPPLLTDDMIAAYTAALVSGTATAGWTFGAGAALQIPFAMQ
AGZ48806.1 GDINARDLICAQKFNGLTVLPPLLTDDMIAAYTAALVSGTATAGWTFGAGAALQIPFAMQ
QDF43825.1 GDINARDLICAQKFNGLTVLPPLLTDDMIAAYTAALVSGTATAGWTFGAGAALQIPFAMQ
AGZ48818.1 GDINARDLICAQKFNGLTVLPPLLTDDMIAAYTAALVSGTATAGWTFGAGAALQIPFAMQ
QHD43416.1 GDIAARDLICAQKFNGLTVLPPLLTDEMIAQYTSALLAGTITSGWTFGAGAALQIPFAMQ
AVP78031.1 GGISARDLICAQKFNGLTVLPPLLTDEMIAAYTAALISGTATAGWTFGAGAALQIPFAMQ
ABD75323.1 GGINARDLICAQKFNGLTVLPPLLTDDMIAAYTAALISGTATAGWTFGAGAALQIPFAMQ
QDF43835.1 GDINARDLICAQKFNGLTVLPPLLTDDMIAAYTAALVSGTATAGWTFGAGAALQIPFAMQ
ABD75332.1 GDISARDLICAQKFNGLTVLPPLLTDEMIAAYTAALVSGTATAGWTFGAGSALQIPFAMQ
QDF43820.1 GDINARDLICAQKFNGLTVLPPLLTDDMIAAYTAALVSGTATAGWTFGAGAALQIPFAMQ
AAZ67052.1 GDISARDLICAQKFNGLTVLPPLLTDEMIAAYTAALVSGTATAGWTFGAGSALQIPFAMQ
AFS88936.1 GPASARDLICAQYVAGYKVLPPLMDVNMEAAYTSSLLGSIAGVGWTAGLSSFAAIPFAQS
YP_0010399 GPQSARDLICAQYVSGYKVLPPLYDPNMEAAYTSSLLGSIAGAGWTAGLSSFAAIPFAQS
* ******** . * .***** :* * **::*:.. *** * .: **** .
ALK02457.1 MAYRFNGIGVTQNVLYENQKQIANQFNKAISQIQESLTTTSTALGKLQDVVNQNAQALNT
AAS10463.1 MAYRFNGIGVTQNVLYENQKQIANQFNKAISQIQESLTTTSTALGKLQDVVNQNAQALNT
AAP13441.1 MAYRFNGIGVTQNVLYENQKQIANQFNKAISQIQESLTTTSTALGKLQDVVNQNAQALNT
AAP13567.1 MAYRFNGIGVTQNVLYENQKQIANQFNKAISQIQESLTTTSTALGKLQDVVNQNAQALNT
AGZ48806.1 MAYRFNGIGVTQNVLYENQKQIANQFNKAISQIQESLTTTSTALGKLQDVVNQNAQALNT
QDF43825.1 MAYRFNGIGVTQNVLYENQKQIANQFNKAISQIQESLTTTSTALGKLQDVVNQNAQALNT
AGZ48818.1 MAYRFNGIGVTQNVLYENQKQIANQFNKAISQIQESLTTTSTALGKLQDVVNQNAQALNT
QHD43416.1 MAYRFNGIGVTQNVLYENQKLIANQFNSAIGKIQDSLSSTASALGKLQDVVNQNAQALNT
AVP78031.1 MAYRFNGIGVTQNVLYENQKLIANQFNSAIGKIQESLTSTASALGKLQDVVNQNAQALNT
ABD75323.1 MAYRFNGIGVTQNVLYENQKQIANQFNKAITQIQESLTTTSTALGKLQDVVNQNAQALNT
QDF43835.1 MAYRFNGIGVTQNVLYENQKQIANQFNKAISQIQESLTTTSTALGKLQDVVNQNAQALNT
ABD75332.1 MAYRFNGIGVTQNVLYENQKQIANQFNKAISQIQESLTTTSTALGKLQDVVNQNAQALNT
QDF43820.1 MAYRFNGIGVTQNVLYENQKQIANQFNKAISQIQESLTTTSTALGKLQDVVNQNAQALNT
AAZ67052.1 MAYRFNGIGVTQNVLYENQKQIANQFNKAISQIQESLTTTSTALGKLQDVVNQNAQALNT
AFS88936.1 IFYRLNGVGITQQVLSENQKLIANKFNQALGAMQTGFTTTNEAFQKVQDAVNNNAQALSK
YP_0010399 MFYRLNGVGITQQVLSENQKLIANKFNQALGAMQTGFTTSNLAFSKVQDAVNANAQALSK
: **:**:*:**:** **** ***:**.*: :* .:::: *: *:**.** *****..
ALK02457.1 LVKQLSSNFGAISSVLNDILSRLDKVEAEVQIDRLITGRLQSLQTYVTQQLIRAAEIRAS
AAS10463.1 LVKQLSSNFGAISSVLNDILSRLDKVEAEVQIDRLITGRLQSLQTYVTQQLIRAAEIRAS
AAP13441.1 LVKQLSSNFGAISSVLNDILSRLDKVEAEVQIDRLITGRLQSLQTYVTQQLIRAAEIRAS
AAP13567.1 LVKQLSSNFGAISSVLNDILSRLDKVEAEVQIDRLITGRLQSLQTYVTQQLIRAAEIRAS
AGZ48806.1 LVKQLSSNFGAISSVLNDILSRLDKVEAEVQIDRLITGRLQSLQTYVTQQLIRAAEIRAS
QDF43825.1 LVKQLSSNFGAISSVLNDILSRLDKVEAEVQIDRLITGRLQSLQTYVTQQLIRAAEIRAS
AGZ48818.1 LVKQLSSNFGAISSVLNDILSRLDKVEAEVQIDRLITGRLQSLQTYVTQQLIRAAEIRAS
QHD43416.1 LVKQLSSNFGAISSVLNDILSRLDKVEAEVQIDRLITGRLQSLQTYVTQQLIRAAEIRAS
AVP78031.1 LVKQLSSNFGAISSVLNDILSRLDKVEAEVQIDRLITGRLQSLQTYVTQQLIRAAEIRAS
ABD75323.1 LVKQLSSNFGAISSALNDILSRLDKVEAEVQIDRLITGRLQSLQTYVTQQLIRAAEIRAS
QDF43835.1 LVKQLSSNFGAISSVLNDILSRLDKVEAEVQIDRLITGRLQSLQTYVTQQLIRAAEIRAS
ABD75332.1 LVKQLSSNFGAISSVLNDILSRLDKVEAEVQIDRLITGRLQSLQTYVTQQLIRAAEIRAS
QDF43820.1 LVKQLSSNFGAISSVLNDILSRLDKVEAEVQIDRLITGRLQSLQTYVTQQLIRAAEIRAS
AAZ67052.1 LVKQLSSNFGAISSVLNDILSRLDKVEAEVQIDRLITGRLQSLQTYVTQQLIRAAEIRAS
AFS88936.1 LASELSNTFGAISASIGDIIQRLDVLEQDAQIDRLINGRLTTLNAFVAQQLVRSESAALS
YP_0010399 LASELSNTFGAISSSISDILARLDTVEQDAQIDRLINGRLISLNAFVSQQLVRSETAARS
*..:**..*****: :.**: *** :* :.******.*** :*:::*:***:*: *
ALK02457.1 ANLAATKMSECVLGQSKRVDFCGKGYHLMSFPQAAPHGVVFLHVTYVPSQERNFTTAPAI
AAS10463.1 ANLAATKMSECVLGQSKRVDFCGKGYHLMSFPQAAPHGVVFLHVTYVPSQERNFTTAPAI
AAP13441.1 ANLAATKMSECVLGQSKRVDFCGKGYHLMSFPQAAPHGVVFLHVTYVPSQERNFTTAPAI
AAP13567.1 ANLAATKMSECVLGQSKRVDFCGKGYHLMSFPQAAPHGVVFLHVTYVPSQERNFTTAPAI
AGZ48806.1 ANLAATKMSECVLGQSKRVDFCGKGYHLMSFPQAAPHGVVFLHVTYVPSQERNFTTAPAI
QDF43825.1 ANLAATKMSECVLGQSKRVDFCGKGYHLMSFPQAAPHGVVFLHVTYVPSQERNFTTAPAI
AGZ48818.1 ANLAATKMSECVLGQSKRVDFCGKGYHLMSFPQAAPHGVVFLHVTYVPSQERNFTTAPAI
QHD43416.1 ANLAATKMSECVLGQSKRVDFCGKGYHLMSFPQSAPHGVVFLHVTYVPAQEKNFTTAPAI
AVP78031.1 ANLAATKMSECVLGQSKRVDFCGKGYHLMSFPQSAPHGVVFLHVTYIPSQEKNFTTAPAI
ABD75323.1 ANLAATKMSECVLGQSKRVDFCGKGYHLMSFPQSAPHGVVFLHVTYVPSQEKNFTTAPAI
QDF43835.1 ANLAATKMSECVLGQSKRVDFCGRGYHLMSFPQAAPHGVVFLHVTYVPSQEKNFTTAPAI
ABD75332.1 ANLAATKMSECVLGQSKRVDFCGKGYHLMSFPQAAPHGVVFLHVTYVPSQERNFTTAPAI
QDF43820.1 ANLAATKMSECVLGQSKRVDFCGRGYHLMSFPQAAPHGVVFLHVTYVPSQEKNFTTAPAI
AAZ67052.1 ANLAATKMSECVLGQSKRVDFCGKGYHLMSFPQAAPHGVVFLHVTYVPSQERNFTTAPAI
AFS88936.1 AQLAKDKVNECVKAQSKRSGFCGQGTHIVSFVVNAPNGLYFMHVGYYPSNHIEVVSAYGL
YP_0010399 AQLASDKVNECVKSQSKRNGFCGSGTHIVSFVVNAPNGFYFFHVGYVPTNYTNVTAAYGL
*:** *:.*** .**** .*** * *::** **:*. *:** * *:: :..:* .:
ALK02457.1 CHEGK---AYFPREGVFVFNGTS-------WFITQRNFFSPQIITTDNT-FVSGSCDVVI
AAS10463.1 CHEGK---AYFPREGVFVFNGTS-------WFITQRNFFSPQIITTDNT-FVSGNCDVVI
AAP13441.1 CHEGK---AYFPREGVFVFNGTS-------WFITQRNFFSPQIITTDNT-FVSGNCDVVI
AAP13567.1 CHEGK---AYFPREGVFVFNGTS-------WFITQRNFFSPQIITTDNT-FVSGNCDVVI
AGZ48806.1 CHEGK---AYFPREGVFVFNGTS-------WFITQRNFFSPQIITTDNT-FVSGSCDVVI
QDF43825.1 CHEGK---AYFPREGVFVFNGTS-------WFITQRNFFSPQIITTDNT-FVSGSCDVVI
AGZ48818.1 CHEGK---AYFPREGVFVFNGTS-------WFITQRNFFSPQIITTDNT-FVSGSCDVVI
QHD43416.1 CHDGK---AHFPREGVFVSNGTH-------WFVTQRNFYEPQIITTDNT-FVSGNCDVVI
AVP78031.1 CHEGK---AHFPREGVFVSNGTH-------WFVTQRNFYEPKIITTDNT-FVSGNCDVVI
ABD75323.1 CHEGK---AYFPREGVFVSNGSS-------WFITQRNFYSPQIITTDNT-FVAGSCDVVI
QDF43835.1 CHEGK---AYFPREGVFVSNGTS-------WFITQRNFYSPQIITTDNT-FVAGSCDVVI
ABD75332.1 CHEGK---AYFPREGVFVSNGTS-------WFITQRNFYSPQIITTDNT-FVAGNCDVVI
QDF43820.1 CHEGK---AYFPREGVFVSNGTF-------WFITQRNFYSPQIITTDNT-FVAGNCDVVI
AAZ67052.1 CHEGK---AYFPREGVFVSNGTS-------WFITQRNFYSPQIITTDNT-FVAGSCDVVI
AFS88936.1 CDAANPTNCIAPVNGYFIKTNNT--RIVDEWSYTGSSFYAPEPITSLNTKYVA--PQVTY
YP_0010399 CNNNNPPLCIAPIDGYFITNQTTTYSVDTEWYYTGSSFYKPEPITQANSRYVS--SDVKF
* : . * :* *: . . * * .*: *: ** *: :*: :*
ALK02457.1 GIINNTVYDPL---QPELDSFKEELDKYFKNHTSPDVDLGDISGINASVVNIQKEIDRLN
AAS10463.1 GIINNTVYDPL---QPELDSFKEELDKYFKNHTSPDVDLGDISGINASVVNIQEEIDRLN
AAP13441.1 GIINNTVYDPL---QPELDSFKEELDKYFKNHTSPDVDLGDISGINASVVNIQKEIDRLN
AAP13567.1 GIINNTVYDPL---QPELDSFKEELDKYFKNHTSPDVDLGDISGINASVVNIQKEIDRLN
AGZ48806.1 GIINNTVYDPL---QPELDSFKEELDKYFKNHTSPDVDLGDISGINASVVNIQKEIDRLN
QDF43825.1 GIINNTVYDPL---QPELDSFKEELDKYFKNHTSPDVDLGDISGINASVVNIQKEIDRLN
AGZ48818.1 GIINNTVYDPL---QPELDSFKEELDKYFKNHTSPDVDLGDISGINASVVNIQKEINRLN
QHD43416.1 GIVNNTVYDPL---QPELDSFKEELDKYFKNHTSPDVDLGDISGINASVVNIQKEIDRLN
AVP78031.1 GIINNTVYDPL---QPELDSFKEELDKYFKNHTSPDIDLGDISGINASVVNIQKEIDRLN
ABD75323.1 GIINNTVYDPL---QPELDSFKQELDKYFKNHTSPDVDLGDISGINASVVDIQKEIDRLN
QDF43835.1 GIINNTVYDPL---QPELDSFKEELDKYFKNHTSPDVDLGDISGINASVVNIQKEIDRLN
ABD75332.1 GIINNTVYDPL---QPELDSFKEELDKYFKNHTSPDVDLGDISGINASVVNIQKEIDRLN
QDF43820.1 GIINNTVYDPL---QPELDSFKEELDKYFKNHTSPDVDLGDISGINASVVNIQKEIDRLN
AAZ67052.1 GIINNTVYDPL---QPELDSFKEELDKYFKNHTSPDVDLGDISGINASVVNIQKEIDRLN
AFS88936.1 QNISTNLPPPLLGNSTGID-FQDELDEFFKNVSTSIPNFGSLTQINTTLLDLTYEMLSLQ
YP_0010399 DKLENNLPPPLLENSTDVD-FKDELEEFFKNVTSHGPNFAEISKINTTLLDLSDEMAMLQ
:...: ** .. :* *::**:::*** :: ::..:: **:::::: *: *:
ALK02457.1 EVAKNLNESLIDLQELGKYEQYIKWPWYVWLGFIAGLIAIVMVTILLCCMTSCCSCLKGA
AAS10463.1 EVAKNLNESLIDLQELGKYEQYIKWPWYVWLGFIAGLIAIVMVTILLCCMTSCCSCLKGA
AAP13441.1 EVAKNLNESLIDLQELGKYEQYIKWPWYVWLGFIAGLIAIVMVTILLCCMTSCCSCLKGA
AAP13567.1 EVAKNLNESLIDLQELGKYEQYIKWPWYVWLGFIAGLIAIVMVTILLCCMTSCCSCLKGA
AGZ48806.1 EVAKNLNESLIDLQELGKYEQYIKWPWYVWLGFIAGLIAIVMVTILLCCMTSCCSCLKGA
QDF43825.1 EVAKNLNESLIDLQELGKYEQYIKWPWYVWLGFIAGLIAIVMVTILLCCMTSCCSCLKGA
AGZ48818.1 EVAKNLNESLIDLQELGKYEQYIKWPWYVWLGFIAGLIAIVMVTILLCCMTSCCSCLKGA
QHD43416.1 EVAKNLNESLIDLQELGKYEQYIKWPWYIWLGFIAGLIAIVMVTIMLCCMTSCCSCLKGC
AVP78031.1 EVARNLNESLIDLQELGKYEQYIKWPWYVWLGFIAGLIAIVMVTILLCCMTSCCSCLKGC
ABD75323.1 EVAKNLNESLIDLQELGKYEQYIKWPWYVWLGFIAGLVGLFMAIILLCYFTSCCSCCKGM
QDF43835.1 EVAKNLNESLIDLQELGKYEQYIKWPWYVWLGFIAGLIAIVMATILLCCMTSCCSCLKGA
ABD75332.1 EVAKNLNESLIDLQELGKYEQYIKWPWYVWLGFIAGLIAIVMVTILLCCMTSCCSCLKGA
QDF43820.1 EVAKNLNESLIDLQELGKYEQYIKWPWYVWLGFIAGLIAIVMATILLCCMTSCCSCLKGA
AAZ67052.1 EVAKNLNESLIDLQELGKYEQYIKWPWYVWLGFIAGLIAIVMVTILLCCMTSCCSCLKGA
AFS88936.1 QVVKALNESYIDLKELGNYTYYNKWPWYIWLGFIAGLVALALCVFFILCCTGCGTNCMGK
YP_0010399 EVVKQLNDSYIDLKELGNYTYYNKWPWYVWLGFIAGLVALLLCVFFLLCCTGCGTSCLGK
:*.. **:* ***:***:* * *****:********:.: : ::: *.* : *
ALK02457.1 CSCGSCC-KFDEDDSEPVLKGVKLHYT
AAS10463.1 CSCGSCC-KFDEDDSEPVLKGVKLHYT
AAP13441.1 CSCGSCC-KFDEDDSEPVLKGVKLHYT
AAP13567.1 CSCGSCC-KFDEDDSEPVLKGVKLHYT
AGZ48806.1 CSCGSCC-KFDEDDSEPVLKGVKLHYT
QDF43825.1 CSCGSCC-KFDEDDSEPVLKGVKLHYT
AGZ48818.1 CSCGSCC-KFDEDDSEPVLKGVKLHYT
QHD43416.1 CSCGSCC-KFDEDDSEPVLKGVKLHYT
AVP78031.1 CSCGSCC-KFDEDDSEPVLKGVKLHYT
ABD75323.1 CSCGSCC-RFDEDDSEPVLKGVKLHYT
QDF43835.1 CSCGSCC-KFDEDDSEPVLKGVKLHYT
ABD75332.1 CSCGSCC-KFDEDDSEPVLKGVKLHYT
QDF43820.1 CSCGSCC-KFDEDDSEPVLKGVKLHYT
AAZ67052.1 CSCGSCC-KFDEDDSEPVLKGVKLHYT
AFS88936.1 LKCNRCCDRYEEYDLEP----HKVHVH
YP_0010399 MKCKNCCDSYEEYDVE------KIHVH
.* ** ::* * * *:*
- The “*” represents invariant regions, “:” represents highly conserved regions, “.” for weakly conserved, and a space for not conserved regions
- Click on tab 6.Tree Rendering so the data is reformatted as a phylogeny tree
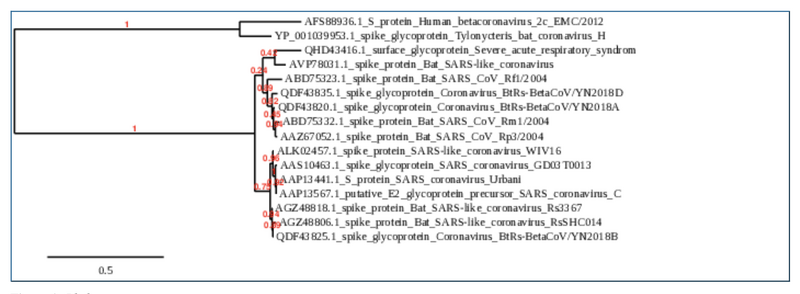
- On the phylogenetic tree, horizontal lines (branches) represents individual evolutionary lineages and vertical lines (splits) represents mutation events
- Comparing the Multiple Sequence Alignment to the Phylogenetic Tree:
- The tree and the sequence alignment both depict highly conserved regions between many of the groups, but specifically between AGZ48806.1 spike protein [Bat SARS-like coronavirus RsSHC014] and AGZ48818.1 spike protein [Bat SARS-like coronavirus Rs3367] there are a few conserved regions within the two sequences that align and on the phylogenetic tree they occupy the same clades sister taxa with a 94% confidence within that clade. This indicates that both sequences are relatively related and share a most common ancestor. Another observation would be the outgroups AFS88936.1 S protein [Human betacoronavirus 2c EMC/2012] and YP_001039953.1 spike glycoprotein [Tylonycteris bat coronavirus HKU4] which are also sister taxa and show the most similar sequences of conservation and variation. When comparing these two groups they depict a lot of variation between one another's conserved regions and variance regions, explaining the distance on the phylogenetic tree.
- Compare the Multiple Sequence alignment to Figure 3 ofWan et al (2020) paper:
- THe class's alignment and the figure were not extremely similar to the figure depicted in the Wan et al. (2020) paper. The amount of invariance in the paper depicted is higher than what was portrayed in the class's sequence alignment. Additionally, the amount of conservation in the paper's figure was significantly higher and did not align completely with areas of conservation within the class's alignment.
- Compare the Multiple Sequence alignment to Figure 2 ofWan et al (2020) paper:
- The sequences within the class did not directly align to those that were present in the figure of the Wan et al. (2020) paper. This resulted I differences being more prevalent than if exact sequences were compared, but the tree, did have the same 2 primary branches, but within clades, did not depict similar trends of outgroups and evolutionary branches.
- Is there adequate enough information to reproduce Wan et al (2020) paper analysis?:
- There is not a sufficient amount of information to completely replicate the results from the Wan et al. (2020) paper. The paper lists an outgroup BtSCoV PDF2386, but the sequences were unavailable on Genbank. Since the exact sequences are unavailable it wouldn't be possible to replicate the exact results of the papers and figures.
Conclusion
The lab compared the sequences of a variety of covid-like and related spike proteins in order to discern the genetic similarities and similarities between multiple species. The purpose of this experiment was to analyze data and build the skills necessary to find species with similarities to the 2019-nCoV in order to better understand the genetic information of 2019-nCoV. The purpose of comparing it to another similar study was to analyze the differences and similarities between our own findings as well as comprehension and application of the knowledge to the information gathered.
Acknowledgements
- I acknowledge my homework partner Taylor Makela, who I spent several hours with going over formatting and general questions about the assignment.
- I acknowledge that I copied and modified the protocol shown on the Week 4 assignment page for this course.
- I acknowledge Anna Horvath who's format for Week 3 I referenced throughout this assignment.
- Except for what is noted above, this individual entry was completed by me and not copied
- Nidapatel (talk) 00:06, 1 October 2020 (PDT)
References
- OpenWetWare. (2020). BIOL368/F20:Week 4. Accessed 30 September 2020, from https://openwetware.org/wiki/BIOL368/F20:Week_4
- OpenWetWare. (2020). Talk:BIOL368/F20:Week 4. Accessed 30 September 2020, from https://openwetware.org/wiki/Talk:BIOL368/F20:Week_4
- NCBI GenBank. (2020). Severe Acute Respiratory Syndrome Coronavirus 2 Isolate Wuhan-Hu-1, Complete Genome. Retrieved 30 September 2020, from https://www.ncbi.nlm.nih.gov/nuccore/MN908947
- NCBI GenBank. (2020). Spike Protein [Bat SARS-like Coronavirus RsSHC014].Accessed 30 September 2020, from https://www.ncbi.nlm.nih.gov/protein/556015117
- Phylogeny.fr: "One Click" Mode. (2020). Accessed 30 September 2020, from http://www.phylogeny.fr/simple_phylogeny.cgi?workflow_id=b9c0813cbbe9695d63cf7e31da5f026d&tab_index=1
- Wan, Y., Shang, J., Graham, R., Baric, R., & Li, F. (2020). Receptor Recognition by the Novel Coronavirus from Wuhan: an Analysis Based on Decade-Long Structural Studies of SARS Coronavirus. Journal Of Virology, 94(7). doi: 10.1128/jvi.00127-20
Template Links
Assignment Pages
- BIOL368/F20:Week 1
- BIOL368/F20:Week 2
- BIOL368/F20:Week 3
- BIOL368/F20:Week 4
- BIOL368/F20:Week 5
- BIOL368/F20:Week 6
- BIOL368/F20:Week 7
- BIOL368/F20:Week 8
- BIOL368/F20:Week 9
- BIOL368/F20:Week 10
- BIOL368/F20:Week 11
- BIOL368/F20:Week 12
- BIOL368/F20:Week 13
- BIOL368/F20:Week 14
Individual Journal Pages
- Nida Patel Journal Week 2
- Nida Patel Journal Week 3
- Nida Patel Journal Week 4
- Nida Patel Journal Week 5
- Nida Patel Journal Week 6
- Nida Patel Journal Week 7
- CancerTracer Review
- Nida Patel Journal Week 10
- Nida Patel Journal Week 11
- Comparison of Human and Hamster ACE2 Receptors for SARS-CoV-2 Week 12
- Nida Patel Journal Week 14
Class Journal Pages
- BIOL368/F20:Class Journal Week 1
- BIOL368/F20:Class Journal Week 2
- BIOL368/F20:Class Journal Week 3
- BIOL368/F20:Class Journal Week 4
- BIOL368/F20:Class Journal Week 5
- BIOL368/F20:Class Journal Week 6
- BIOL368/F20:Class Journal Week 7
- BIOL368/F20:Class Journal Week 8
- BIOL368/F20:Class Journal Week 11
- Comparison of Human and Hamster ACE2 Receptors for SARS-CoV-2 Week 12
- BIOL368/F20:Class Journal Week 13
- BIOL368/F20:Class Journal Week 14